- Title
-
Identification and impact on Pseudomonas aeruginosa virulence of mutations conferring resistance to a phage cocktail for phage therapy
- Authors
- Forti, F., Bertoli, C., Cafora, M., Gilardi, S., Pistocchi, A., Briani, F.
- Source
- Full text @ Microbiol Spectr
|
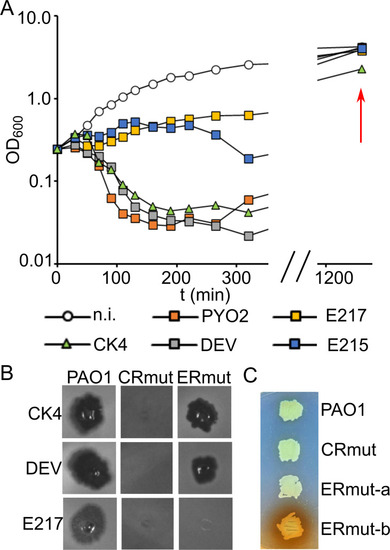
Growth of |
|
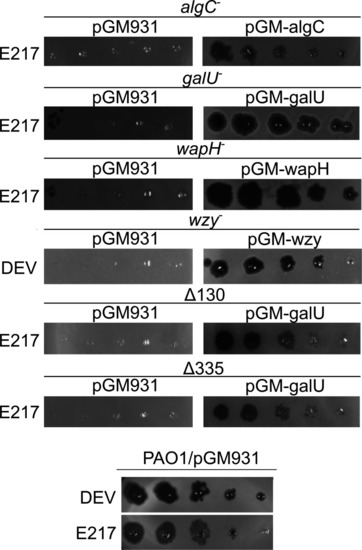
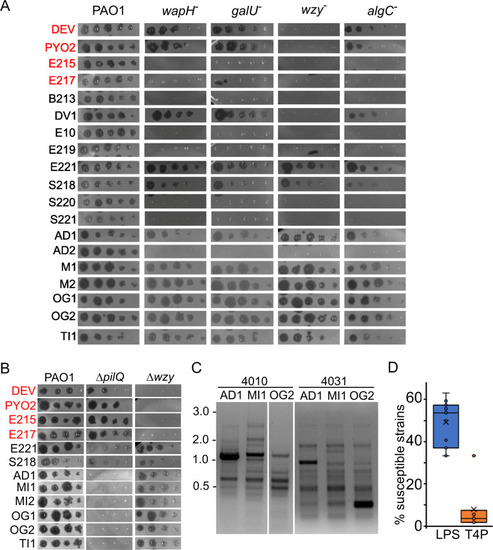
Phage eop on LPS-defective mutants. E217 or DEV lysates (indicated on the left of the panels) were serially diluted (×10) in 96-well plates and replicated as explained in Materials and Methods on PAO1 and PAO1 mutants carrying the indicated plasmids in the presence of 0.2% arabinose in the top agar. The plates were incubated overnight at 37°C. |
|
Altered LPS of phage-resistant mutants and defective adsorption of the cross-resistant |
|
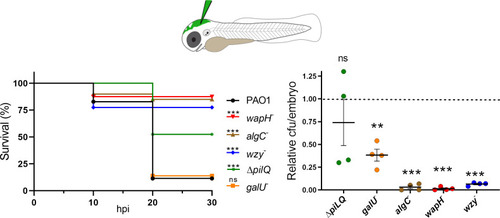
Phage-resistant mutants have attenuated virulence in zebrafish. Left panel. Survival of zebrafish embryos ( PHENOTYPE:
|
|
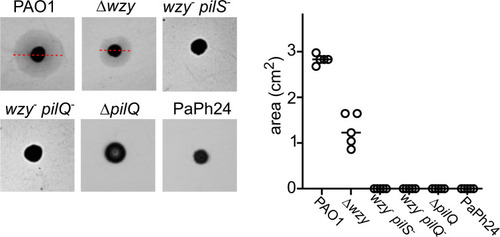
Twitching motility assay. Twitching motility of the indicated strains was assayed as described in Materials and Methods. The results of a representative experiment are shown on the left. The red lines indicate the diameter of the twitching halos formed by PAO1 and Δ |
|
Phage growth on different |