- Title
-
hoxa1a-Null Zebrafish as a Model for Studying HOXA1-Associated Heart Malformation in Bosley-Salih-Alorainy Syndrome
- Authors
- Wang, H., He, J., Han, X., Wu, X., Ye, X., Lv, W., Zu, Y.
- Source
- Full text @ Biology (Basel)
|
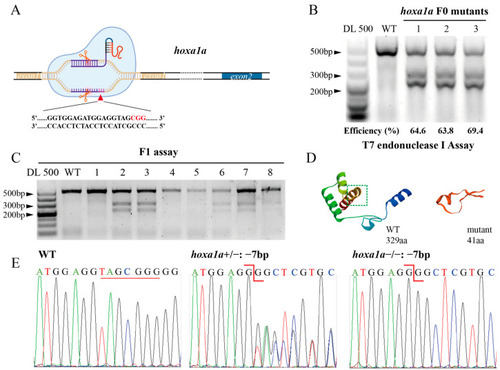
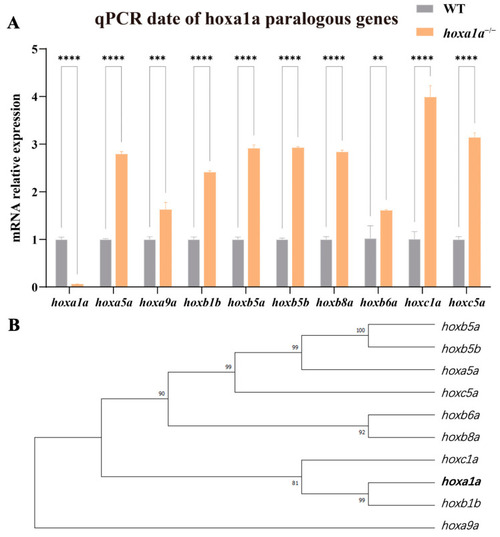
Construction of heritable |
|
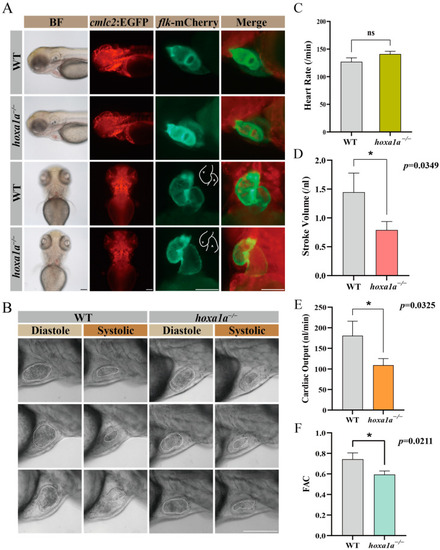
Embryo heart imaging by fluorescence and high-speed camera. ( |
|
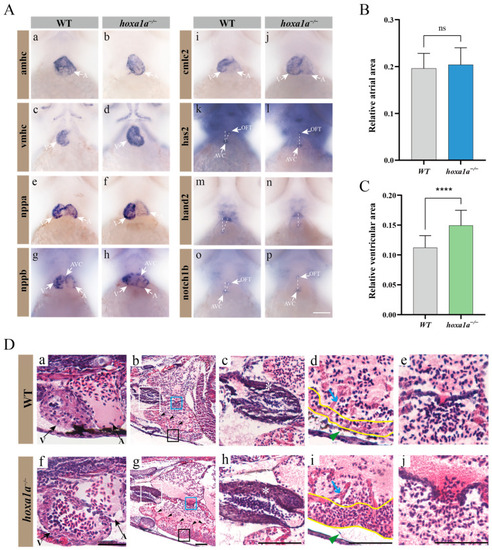
Multiple heart malformation in |
|
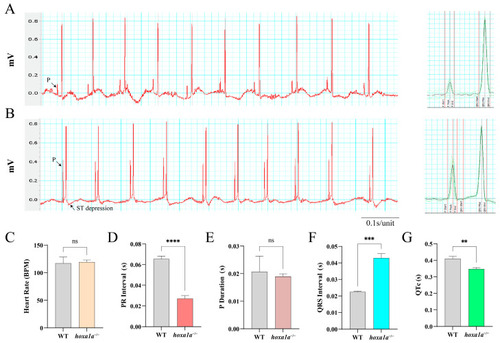
ECG analysis of adult zebrafish. Electrical signal obtained from chest muscle layer in WT ( |
|
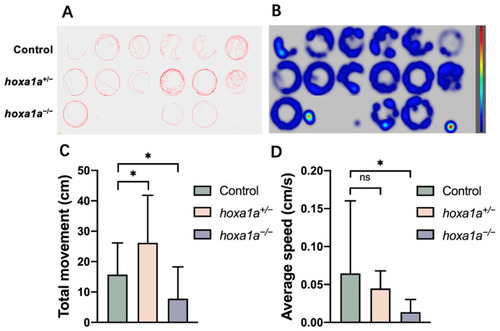
Behavior test for mobility analysis in in |
|
|