- Title
-
Zebrafish as a model for cardiac disease; Cryo-EM structure of native cardiac thin filaments from Danio Rerio
- Authors
- Bradshaw, M., Squire, J.M., Morris, E., Atkinson, G., Richardson, R., Lees, J., Caputo, M., Bigotti, G.M., Paul, D.M.
- Source
- Full text @ J. Muscle Res. Cell Motil.
|
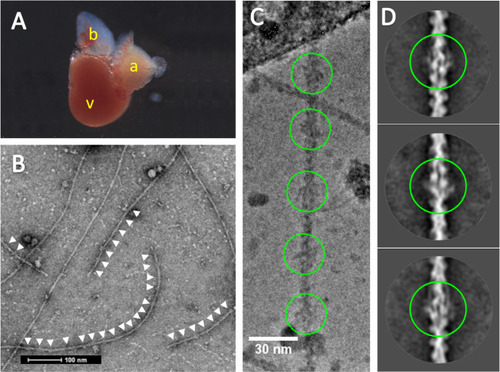
Cryo-EM of zebrafish cardiac thin filaments. |
|
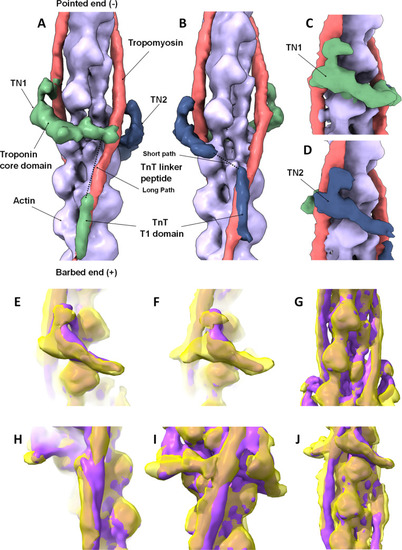
3D reconstruction of native zebrafish cardiac thin filaments and comparison to human reconstituted thin filament structure. |
|
Zebrafish high Ca2+ thin filament model. The full-length actin (grey), TnC (residues 2-161, zebrafish 2-161) green, TnI (41–166, zebrafish 10–135) yellow, TnT (99–272, zebrafish 101–207) orange, full length tropomyosin red, atomic models docked into the electron density. |
|
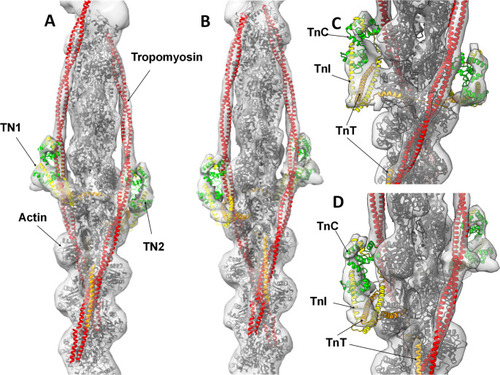
Conformation of troponin core domain and tropomyosin positions. The high and low Ca2+ models 6KN8 (green) and 6KN7 (blue) were docked into our thin filament reconstruction and compared to our zebrafish model (yellow). |
|
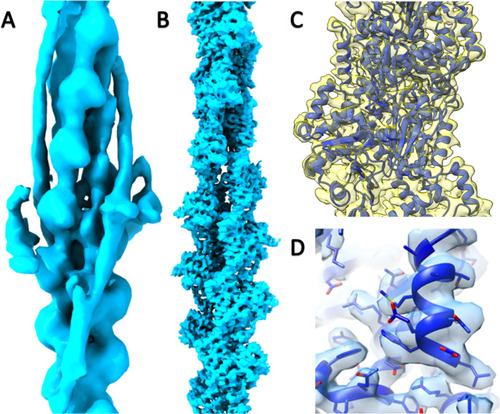
Cryo-EM reconstructions of the zebrafish thin filament and actin filament. |