- Title
-
An in vivo functional assay to characterize human STAT5B genetic variants during zebrafish development
- Authors
- Landi, E., Karabatas, L., Gómez, T.R., Salatino, L., Scaglia, P., Ramírez, L., Keselman, A., Braslavsky, D., Sanguineti, N., Pennisi, P., Rey, R.A., Bergadá, I., Jasper, H.G., Domené, H.M., Plazas, P.V., Domené, S.
- Source
- Full text @ Hum. Mol. Genet.
|
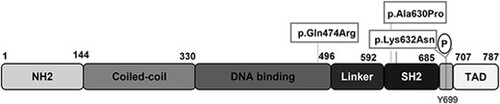
Schematic of the human STAT5b protein adapted from Ramirez et al., 2020. The three variants studied are indicated: p.Gln474Arg located in the DNA binding domain, and p.Ala630Pro and p.Lys632Asn located in the SH2 domain. NH2, amino acids 1?144; Coiled-coil, amino acids 144?330; DNA binding, amino acids 330?496; linker, amino acids 496?592; SH2, amino acids 592?685; TAD, amino acids 707?787. Tyrosine 699 (Y699) can be phosphorylated by JAK2 and other kinases. |
|
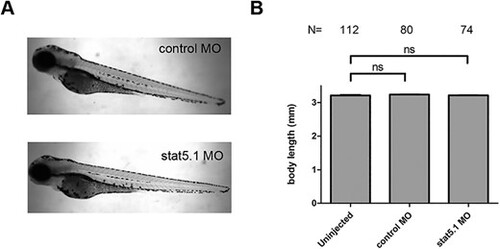
Effect of stat5.1 MO knockdown on zebrafish embryos. (A) Lateral view of representative pictures of control MO and morphant embryos at 72 hpf. (B) Statistical analysis of body length (jaw to tail fin) in uninjected, control MO and stat5.1 MO injected embryos at 72 hpf. Data are shown as mean ± SEM. Mann?Whitney Test. ns = not significant, P > 0.05. N = number of embryos. mm = millimeters. Scale bar: 1 mm. |
|
Effect of overexpression of WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn STAT5B mRNA on zebrafish embryos. (A) Lateral view of representative pictures in uninjected and STAT5B WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn mRNA injected embryos (B) Statistical analysis of body length (jaw to tail fin) in uninjected, and WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn STAT5B mRNA injected embryos. Embryos were measured at 72 hpf. Mann?Whitney test, ***P < 0.0001, ns = not significant, P > 0.05. N = number of embryos. mm = millimeters. Scale bar: 1 mm. |
|
Effect of overexpression of WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn STAT5B mRNA on zebrafish embryos. (A, C, E) Representative pictures of zebrafish embryos with pericardial edema (A), cyclopia (B) and bent spine (E) during development. (B, D, F) Statistical analysis of pericardial edema score (B), cyclopia (D) and bent spine (F) in embryos overexpressing WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn STAT5B mRNA. (G) Percentage of embryos showing pericardial edema, cyclopia, and bent spine in embryos overexpressing WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn STAT5B mRNA. Embryos at 72 hpf were classified into three groups according to the severity of the malformation and scored (1 = normal, 2 = mild and 3 = severe). Mann?Whitney test, ***P < 0.0001, *P < 0.05. N = number of embryos across three independent biological replicates. |
|
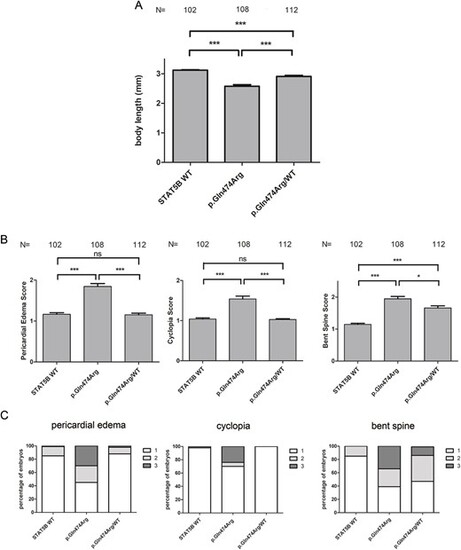
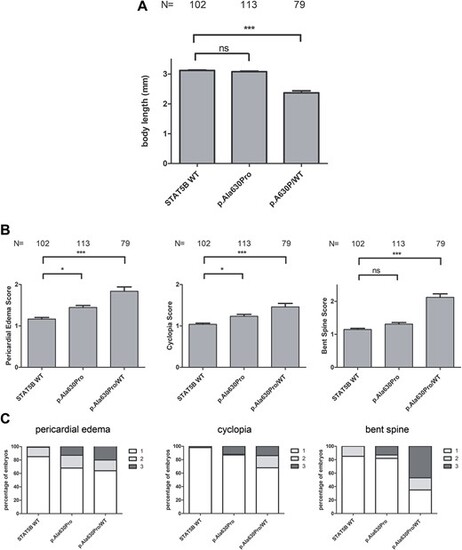
Effect of co-injecting p.Gln474Arg together with WT STAT5B mRNA in a 1:1 ratio in zebrafish embryos. (A) Statistical analysis of body length in embryos overexpressing WT, p.Gln474Arg and p.Gln474Arg/WT STAT5B mRNA. (B) Statistical analysis of pericardial edema, cyclopia and bent spine score in embryos overexpressing WT, p.Gln474Arg and p.Gln474Arg/WT STAT5B mRNA. (C) Percentage of embryos showing pericardial edema, cyclopia and bent spine in embryos overexpressing WT, p.Gln474Arg and p.Gln474Arg/WT STAT5B mRNA. Embryos were scored at 72 hpf and classified into three groups according to the severity of the malformation (1 = normal, 2 = mild, and 3 = severe. Mann?Whitney test, ***P < 0.0001, *P < 0.05, ns = not significant, P > 0.05. N = number of embryos. mm = millimeters. |
|
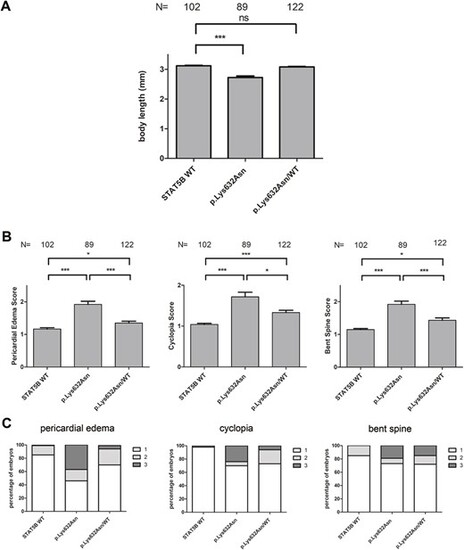
Effect of co-injecting p.Lys632Asn together with WT STAT5B mRNA in a 1:1 ratio in zebrafish embryos. (A) Statistical analysis of body length in embryos overexpressing WT, p.Lys632Asn and p.Lys632Asn/WT STAT5B mRNA. (B) Statistical analysis of pericardial edema, cyclopia and bent spine score in embryos overexpressing WT, p.Lys632Asn and p.Lys632Asn/WT STAT5B mRNA. (C) Percentage of embryos showing pericardial edema, cyclopia and bent spine in embryos overexpressing WT, p.Lys632Asn and p.Lys632Asn/WT STAT5B mRNA. Embryos were scored at 72 hpf and classified into three groups according to the severity of the malformation (1 = normal, 2 = mild and 3 = severe. Mann?Whitney test, ***P < 0.0001, *P < 0.05, ns = not significant, P > 0.05. N = number of embryos. mm = millimeters. |
|
Effect of co-injecting p.Ala630Pro together with WT STAT5B mRNA in a 1:1 ratio in zebrafish embryos. (A) Statistical analysis of body length in embryos overexpressing WT, p.Ala630Pro and p.Ala630Pro/WT STAT5B mRNA. (B) Statistical analysis of pericardial edema, cyclopia and bent spine score in embryos overexpressing WT, p.Ala630Pro and p.Ala630Pro/WT STAT5B mRNA. (C) Percentage of embryos showing pericardial edema, cyclopia and bent spine in embryos overexpressing WT, p.Ala630Pro and p.Ala630Pro/WT STAT5B mRNA. Embryos were scored at 72 hpf and classified into three groups according to the severity of the malformation (1 = normal, 2 = mild and 3 = severe. Mann?Whitney test, ***P < 0.0001, *P < 0.05, ns = not significant, P > 0.05. N = number of embryos. mm = millimeters. |
|
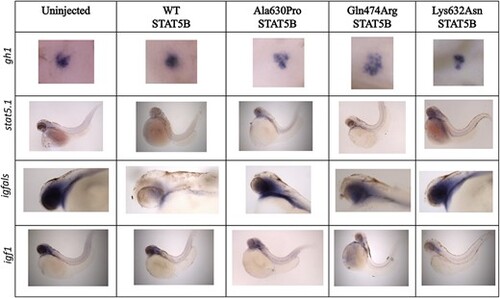
WISH analysis of gh1, stat5.1, igfals and igf1 mRNA in representative zebrafish embryos at 48 hpf. For gh1 expression, embryos are the head region of the embryo, ventral view with anterior to the left. For stat5.1 expression, embryos are whole embryos, lateral view, with anterior to the left and dorsal up. For igfals expression, embryos are the anterior half of the embryo, lateral view with anterior to the left and dorsal up. For igf1 expression, embryos are whole embryos, lateral view, with anterior to the left and dorsal up. Total number of embryos analyzed are shown in Supplementary Material, Table S1. |
|
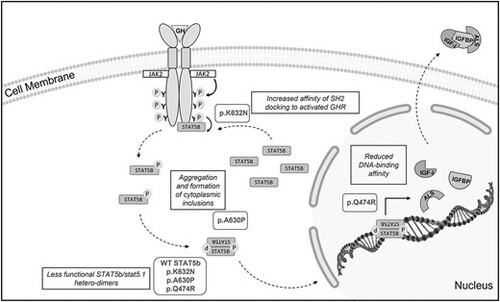
Schematic of the human GH-induced activation of STAT5b. Human STAT5b protein variants p.Ala630Pro, p.Gln474Arg and p.Lys632Asn are shown in bold and the potential mechanisms of interference in the endogenous stat5.1 intracellular signaling cascade are shown in boxes. All variants, including WT, p.Ala630Pro, p.Gln474Arg and p.Lys632Asn STAT5b can interfere with endogenous stat5.1 through the formation of less functional STAT5b/stat5.1 hetero-dimers. In addition, variant p.Ala630Pro can interfere with endogenous stat5.1 by aggregation and through the formation of cytoplasmic inclusions. Variant p.Gln4747Arg can interfere by reduced DNA-binding affinity of the STAT5b/stat5.1 hetero-dimers. Finally, variant p.Lys632Asn can interfere by its increased affinity of SH2 docking to activated GHR. |