- Title
-
GABAA receptor-mediated seizure liabilities: a mixed-methods screening approach
- Authors
- Bampali, K., Koniuszewski, F., Vogel, F.D., Fabjan, J., Andronis, C., Lekka, E., Virvillis, V., Seidel, T., Delaunois, A., Royer, L., Rolf, M.G., Giuliano, C., Traebert, M., Roussignol, G., Fric-Bordat, M., Mazelin-Winum, L., Bryant, S.D., Langer, T., Ernst, M.
- Source
- Full text @ Cell Biol. Toxicol.
|
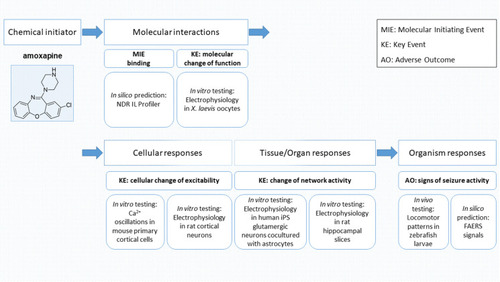
Workflow mapped to adverse outcome pathway (AOP) scales: Assays used for in silico, in vitro, and in vivo experiments to identify molecular initiating events (MIEs) and key events (KEs) related to structural alerts for identifying seizure risk adverse outcomes (AOs) |
|
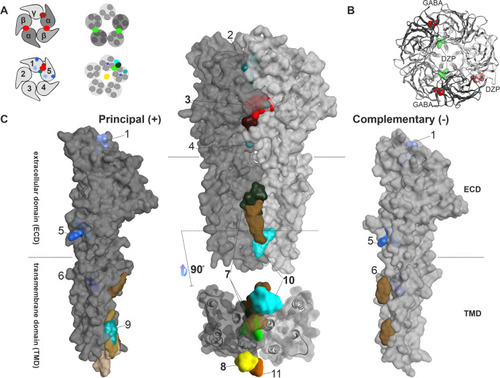
Summary of all identified and putative binding sites, for which structural evidence exists. |
|
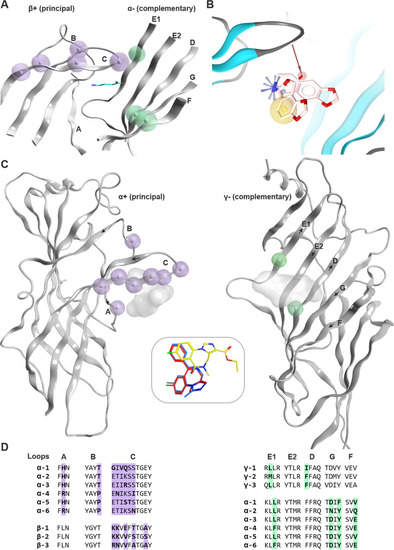
Known ECD interface binding sites for GABA and benzodiazepines: |
|
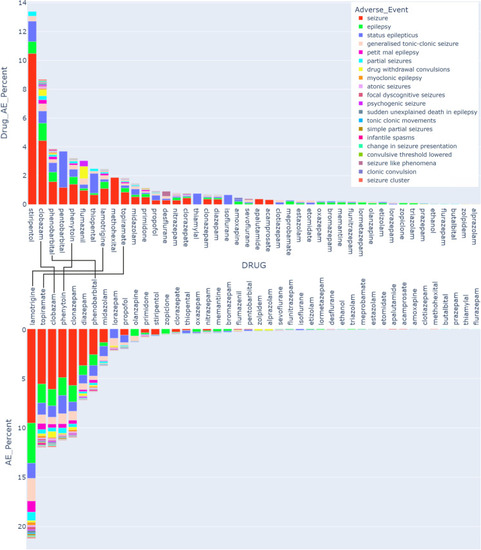
Pharmacovigilance data. The upper box plot displays the fraction (in %) of each AE for a specific drug from the total number of reports for this drug. The drugs are sorted by the total burden of the seizure/convulsion groups of AE, and the AEs in the legend are also sorted by the size of their contribution to the total AE count for these drugs. The lower box plot displays the fraction (in %) of the cumulative seizure AEs for each drug among the total reports in the seizures MedDRA category (46,285 total reports). Compounds which occur in the top 10 of both are connected. The total reports and the reports per AE per drug for the selected drugs and AEs are shown in Supplementary Table |
|
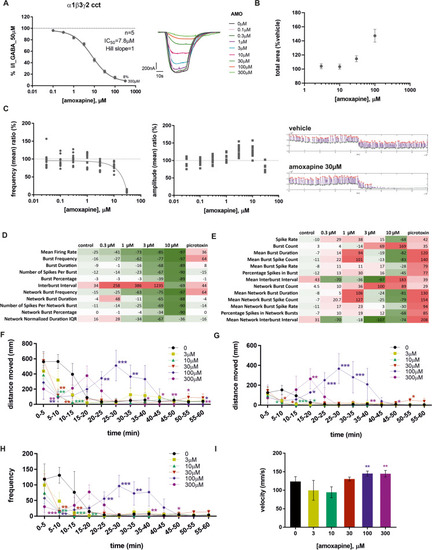
Amoxapine results from different assays. |
|
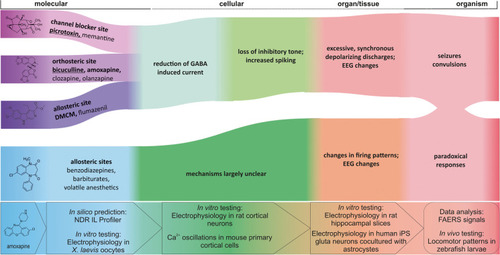
Candidate AOPs for several of the compounds we investigated in this study. The left set of events in blue/ purple hues represent the binding of ligands to their respective binding sites. Picrotoxin, bicuculline, DMCM, clobazam, and amoxapine are rendered in 2D as examples for the different binding sites and for the experimental pipeline. Different allosteric sites can mediate functional agonism and antagonism as well as NAM and PAM effects, which induce typically a change in GABA-elicited current. The proposal reflects a coarse grain model which requires further details to generate complete AOPs. Here, green hues indicate the late molecular and the cellular scales at which the ligand binding leads to changes in inhibitory transmission and then to changes in neuronal firing patterns. The red hues represent the organ and organism scales, at which changed neuronal firing patterns impact on network activity and thus on EEG and ultimately lead to organism responses such as seizures, convulsions, or paradoxical responses such as agitation that are often observed for GABAAR targeting “tranquilizers.” The assays that were used in this study at the molecular, cellular, tissue, and organism scales are integrated at the bottom of the graph. Ligand examples for each pathway are boldfaced and underlined for agents with known seizurogenic properties, boldfaced for strong candidates (meeting at least two criteria), and in standard font for the remaining examples |