- Title
-
Isoalantolactone protects against ethanol-induced gastric ulcer via alleviating inflammation through regulation of PI3K-Akt signaling pathway and Th17 cell differentiation
- Authors
- Zhou, C., Chen, J., Liu, K., Maharajan, K., Zhang, Y., Hou, L., Li, J., Mi, M., Xia, Q.
- Source
- Full text @ Biomed. Pharmacother.
|
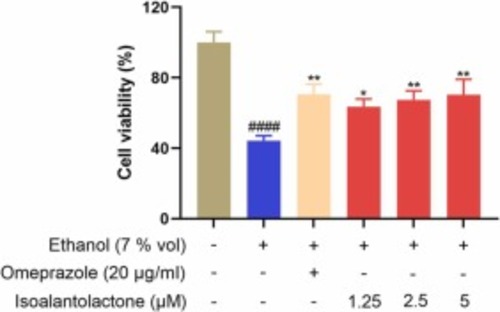
Effects of different concentrations of IAL treatment on cell viability after ethanol injury. Data are expressed as mean ± SEM (n = 5). ####P < 0.0001 vs. the control group, *P < 0.05, **P < 0.01 vs. the model group. |
|
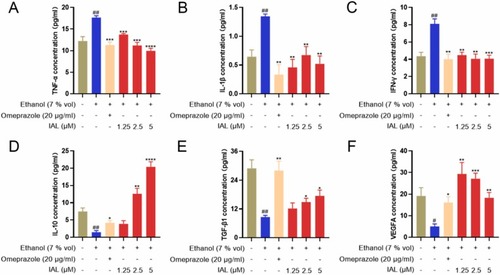
Effect of IAL on the levels of pro-inflammatory cytokines (TNF-α, IL-1β, and IFN-γ) and anti-inflammatory cytokines (IL-10, TGF-β1, and VEGFA), in ethanol-stimulated GES-1 supernatant. Data are expressed as mean ± SEM (n = 5). #P < 0.05, ##P < 0.01 vs. the control group, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. the model group. |
|
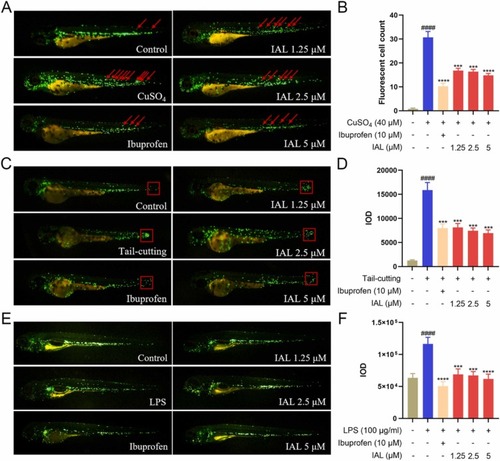
IAL inhibits CuSO4, tail-cutting, and LPS-induced inflammatory responses in zebrafish. (A) Changes of zebrafish immune cell migration in CuSO4-induced inflammatory model. The red arrows indicate inflammatory cells that migrated to the lateral line. (B) The number of inflammatory cells that migrated to the lateral line. (C) Changes of zebrafish immune cell migration in the tail cutting-induced inflammatory model. The red boxes indicate inflammatory cells that accumulate in the tail. (D) The IOD values of immune cells accumulated in the tail. (E) Changes of zebrafish immune cell migration in the LPS-induced inflammatory model. (F) The IOD values of zebrafish systemic immune cells. Data are expressed as mean ± SEM (n = 10). ####P < 0.0001 vs. the control group, ***P < 0.001, ****P < 0.0001 vs. the model group. |
|
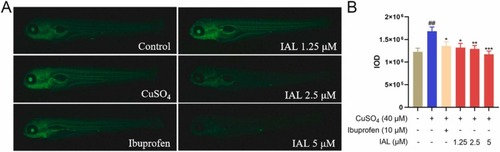
IAL alleviated inflammation in zebrafish larvae by inhibiting CuSO4-induced ROS production. (A) Microphotographs exhibited ROS production in zebrafish larvae. (B) IAL dose-dependently reduced ROS generation induced by CuSO4. Data are expressed as mean ± SEM (n = 10). ##P < 0.01 vs. the control group, *P < 0.05, **P < 0.01, ***P < 0.001 vs. the model group. |
|
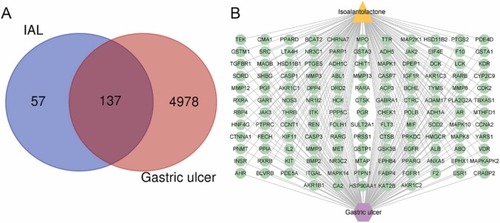
Venn diagram and “IAL-targets-GU” network diagram. (A) Venn diagram of targets between IAL and GU. (B) The “IAL-targets-GU” visualization network diagram. |
|
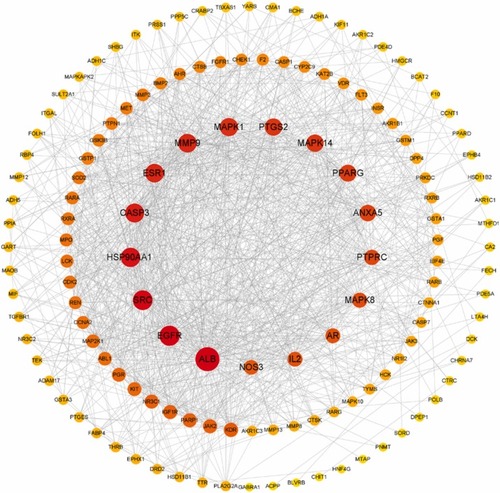
The “protein-protein interaction” (PPI) network diagram. |
|
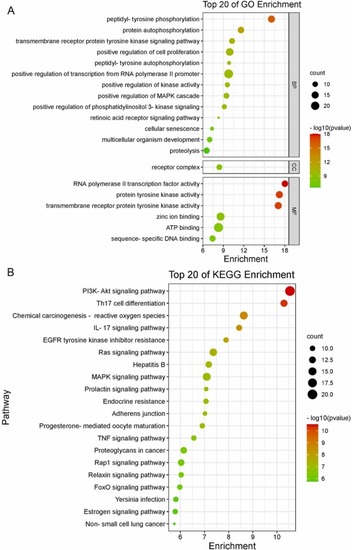
GO and KEGG enrichment analysis. (A) GO enrichment analysis diagram. (B) KEGG enrichment analysis diagram. |
|
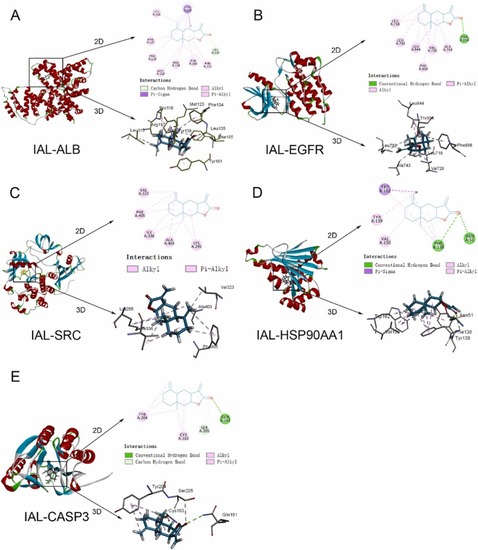
Molecular docking of IAL with 5 targets protein. Interaction of IAL with the target ALB (A), EGFR (B), SRC (C), HSP90AA1 (D), and CASP3 (E). |
|
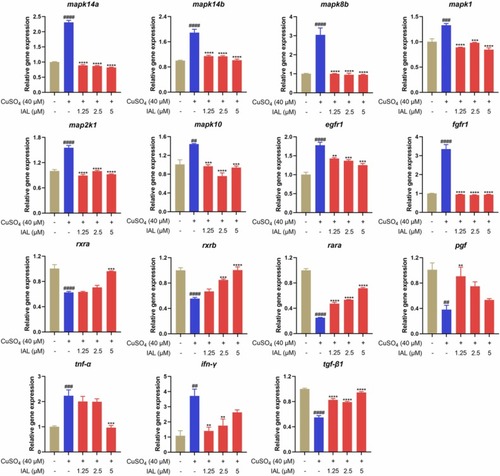
Effect of IAL on the expression levels of inflammation-related genes in zebrafish. Data are expressed as mean ± SEM (n = 3). ##P < 0.01, ###P < 0.001, ####P < 0.0001 vs. the control group, **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. the model group. |