- Title
-
A Drosophila model of the neurological symptoms in Mpv17-related diseases
- Authors
- Kodani, A., Yamaguchi, M., Itoh, R., Huynh, M.A., Yoshida, H.
- Source
- Full text @ Sci. Rep.
|
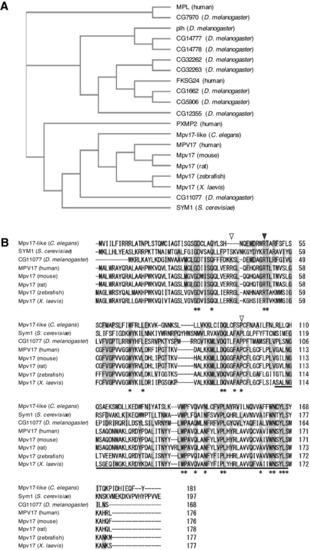
CG11077 is an ortholog of human MPV17. (A) A phylogenetic tree was constructed with amino acid sequences of MPV17/PMP22 family proteins, human (MPL, FKSG24, PXMP22, and MPV17), S. cerevisiae SYM1, D. melanogaster (CG7970, CG14777, CG144778, CG32262, CG32263, CG1662, CG12355, and CG11077), C. elegans Mpv17-like, and Mpv17s (mouse, rat, zebrafish, and X. laevis). The sequence similarity of MPV17/PMP22 family proteins among model organisms and the human MPV17 family was analyzed. (B) Multiple alignment of amino acid sequences of Mpv17 among model organisms and human, including CG11077, was carried out with the Clustal Omega multiple sequence alignment program. Amino acids conserved among all organisms are marked by asterisks and those conserved between D. melanogaster and others are highlighted in gray. The black and white arrowheads indicate the amino acids mutated in MPV17-related hepatocerebral MDDS (R50Q) and CMT (R41Q and P98L) patients, respectively. Underlining indicates the amino acid sequences conserved between PMP22 and MPV17 (Mpv17/PMP22: IPR007248). |
|
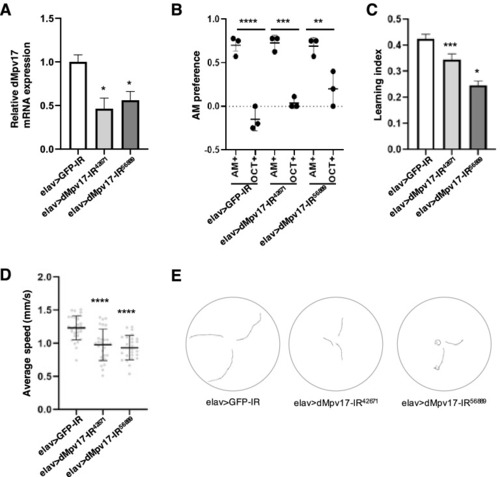
Neuron-specific dMpv17 knockdown larvae show neurological-defect phenotypes. (A) Total RNA was extracted from the CNS of third instar larvae carrying elav > GFP-IR (UAS-GFP-IR/+; elav-GAL4/+;+), elav > dMpv17-IR42671 (elav-GAL4/UAS-dMpv17-IR42671;+), and elav > dMpv17-IR56889 (elav-GAL4/UAS-dMpv17-IR56889;+). The mRNA levels of dMpv17 and G6PD in each strain were detected by qRT-PCR. G6PD was used as an internal control. ***P < 0.01, statistical analysis was performed using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 3. (B, C) The learning ability of larvae carrying elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 was tested in the odor-taste learning assay. AM preference (B) and larval learning index (C) were quantified. *P < 0.05, ***P < 0.001, using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 3. (D, E) Larval locomotor ability was tested in the crawling assay with elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889. (D) The average crawling speed was quantified. Statistical analysis was performed using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 25?31. (E) Typical trajectories of larvae with each genotype. |
|
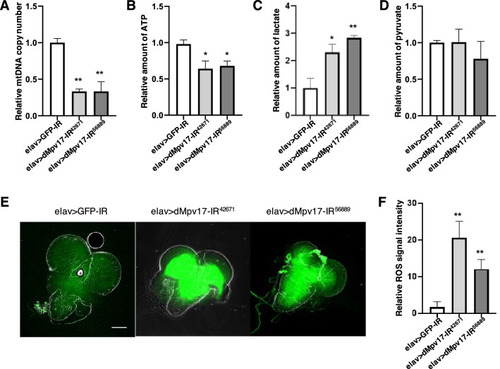
A dMpv17 knockdown induced mitochondrial dysfunction in the larval CNS. (A) The relative mtDNA levels in the larval CNS extract of elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 larvae were quantified. The relative levels of 16s rRNA (for mtDNA) and RpL32 (for nuclear DNA) were measured by qRT-PCR. **P < 0.01, statistical analysis was performed using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 3. (B) The ATP levels in the larval CNS extracts of elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 larvae were measured with Cell titer-Glo assay. *P < 0.05, statistical analysis was performed using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 5. (C) The lactate level in the larval CNS extract of elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 larvae were measured with the Lactate-Glo assay. **P < 0.01, *P < 0.05, statistical analysis was performed using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 3. (D) Pyruvate levels in the CNS extracts of elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 larvae were measured with a pyruvate assay kit. **P < 0.01, *P < 0.05, statistical analysis was performed using one-way ANOVA followed by Dunnett?s multiple comparisons test vs elav > GFP-IR. n = 3. (E) Third instar larval CNSs were stained with CM-H2DCFCA for the detection of ROS. Representative ROS images of the CNS of elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 are shown. Scale bar, 100 ?m. (F) The fluorescence intensity in the CNS area enclosed by the white line was quantified. **P < 0.01, statistical analysis was performed using Student?s t-test vs elav > GFP-IR. n = 5. |
|
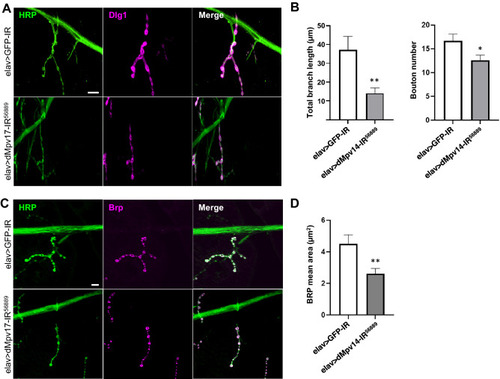
A dMpv17 knockdown influences larval NMJ morphology. (A) Synapses in the NMJ on the fourth muscle of third-instar larvae were inspected in elav > GFP-IR, elav > dMpv17-IR42671, and elav > dMpv17-IR56889 larvae. The presynaptic terminals were visualized with anti-HRP IgG and the post-synaptic domains with anti-Dlg1 IgG. Scale bar, 10 ?m. (B) Synapse branch length, bouton numbers were quantified. **P < 0.01, *P < 0.05, statistical analysis was performed using Student?s t-test vs elav > GFP-IR. n = 15. (C) The active zone in the NMJ of the fourth muscle was visualized with anti-HRP and anti-Brp IgGs. Scale bar, 10 ?m. (D) The area of Brp signals merged with HRP was quantified. **P < 0.01, statistical analysis was performed using Student?s t-test vs elav > GFP-IR. n = 13. |