- Title
-
ST3GAL5-catalyzed gangliosides inhibit TGF-β-induced epithelial-mesenchymal transition via TβRI degradation
- Authors
- Zhang, J., van der Zon, G., Ma, J., Mei, H., Cabukusta, B., Agaser, C.C., Madunić, K., Wuhrer, M., Zhang, T., Ten Dijke, P.
- Source
- Full text @ EMBO J.
|
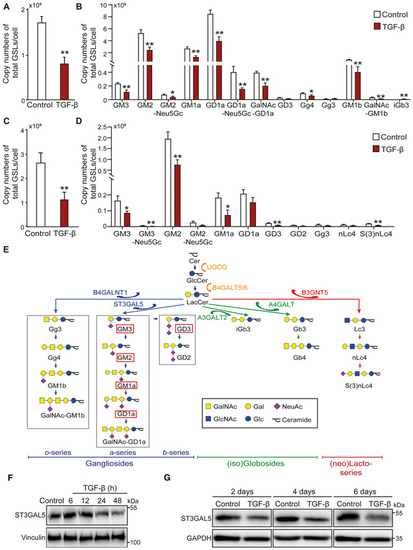
TGF‐β decreases the abundance of glycosphingolipid (GSL)‐glycans
Data information: Unless stated otherwise, TGF‐β (2.5 ng/ml) and vehicle control were applied for 48 h. The histograms show the mean ± SD values from three biological replicates (n = 3). *P ≤ 0.05; **P < 0.01 based on unpaired Student's t‐test. GT1b: internal normalization control. |
|
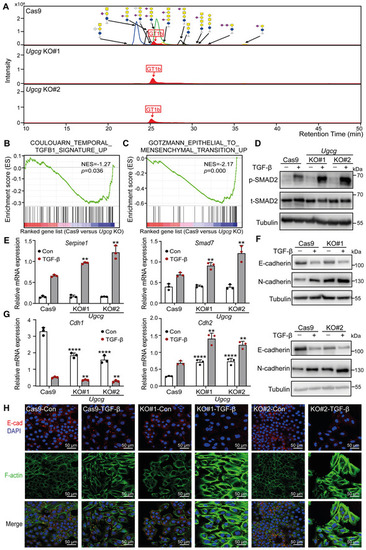
KO of Ugcg, a key enzyme in the biosynthesis of GSLs, promotes TGF‐β signaling and TGF‐β‐induced EMT in NMuMG cells
Data information: TGF‐β was applied at a final concentration of 2.5 ng/ml. In (E, G), the data are expressed as the mean ± SD values from three biological replicates (n = 3). **P < 0.01; ****P < 0.0001 based on unpaired Student's t‐test. |
|
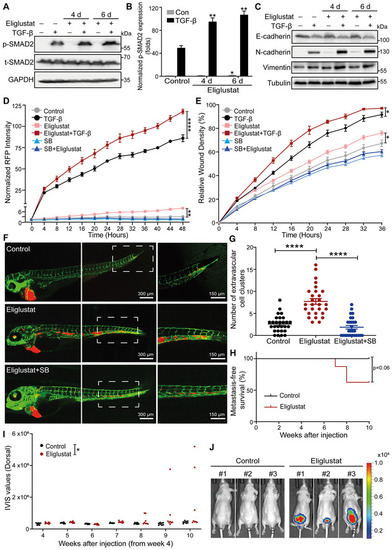
Eliglustat, a UGCG inhibitor, enhances TGF‐β signaling and TGF‐β‐induced EMT, migration, extravasation and early metastatic outgrowth of A549 cells
Data information: TGF‐β and eliglustat were applied at final concentrations of 2.5 ng/ml and 2 μM, respectively. All data are expressed as the mean ± SD values from three biological replicates (n = 3). *P ≤ 0.05; **P < 0.01; ****P < 0.0001. In (B, D, E), statistical analysis was based on two‐way ANOVA. |
|
Inhibition of GSL biosynthesis decreases the TβRI level in lipid rafts and inhibits the ubiquitination and degradation of TβRI
Data information: In (B, D), the data are expressed as the mean ± SD values from two biological replicates (n = 2). In (G), the data are expressed as the means ± SD values from three biological replicates (n = 3). *P ≤ 0.05; **P < 0.01; ****P < 0.0001 based on unpaired Student's t‐test. |
|
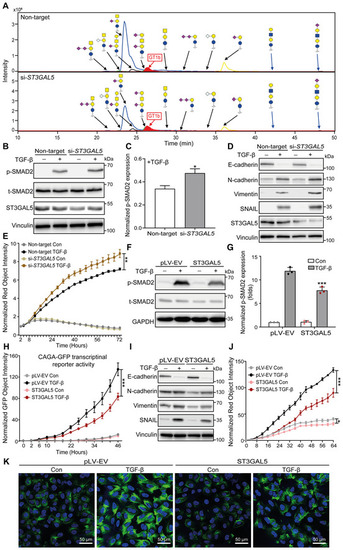
ST3GAL5 inhibits TGF‐β signaling and TGF‐β‐induced EMT
Data information: TGF‐β was applied at a final concentration of 2.5 ng/ml. In (C, E, G, H, J), the data are expressed as the mean ± SD values from three biological replicates (n = 3). *P ≤ 0.05; **P < 0.01; ***P < 0.001. In (C), statistical analysis was based on unpaired Student's t‐test. In (E, G, H, J), statistical analysis was based on two‐way ANOVA. |
|
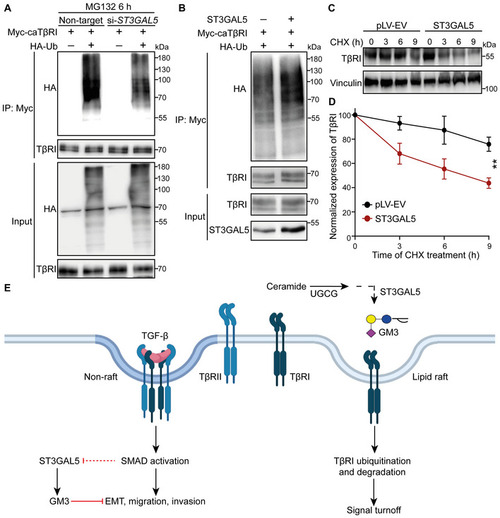
ST3GAL5 promotes TβRI ubiquitination and degradation
|
|
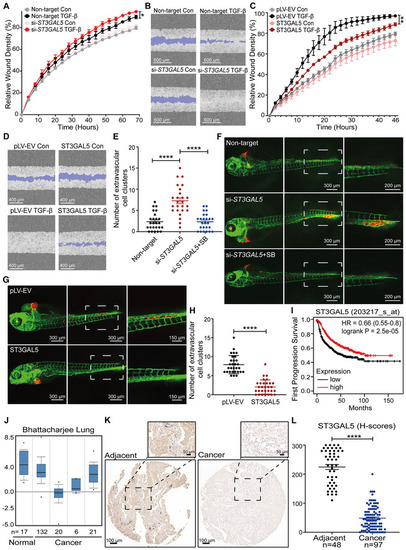
ST3GAL5 inhibits A549 cell migration and invasion, and its expression is associated with good prognosis in lung cancer
|