- Title
-
Urotensin II-related peptides, Urp1 and Urp2, control zebrafish spine morphology
- Authors
- Bearce, E.A., Irons, Z.H., O'Hara-Smith, J.R., Kuhns, C.J., Fisher, S.I., Crow, W.E., Grimes, D.T.
- Source
- Full text @ Elife
|
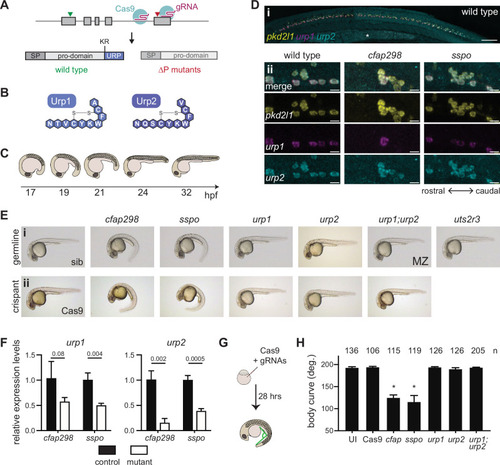
(A) urp1 and urp2 are 5-exon genes (gray boxes). The final exon codes for the 10-amino acid peptides produced after cleavage from the prodomain at a dibasic site (KR). Pairs of gRNAs were used to induce deletions of Urp1 and Urp2 peptide coding sequences, resulting in urp1∆P and urp2∆P mutants, respectively. SP – signal peptide. (B) Urp1 and Urp2 peptide sequences with identical hexacyclic regions. (C) Zebrafish posterior axial straightening, the morphogenetic process which straightens the embryonic body. (D) Fluorescence in situ hybridization based on hybridization chain reaction analysis of pkd2l1, urp1, and urp2 expression in the central canal at 28 hpf. pkd2l1 expression marks CSF-cNs. urp1 expression is restricted to ventral CSF-cNs while urp2 is expressed in all CSF-cNs. Both urp1 and urp2 are expressed in cfap298tm304 and sspob1446 mutants, though comparison of expression between samples was non-quantitative. (i) Shows the zebrafish trunk with the yolk stalk labeled (*). (ii) Shows zoomed regions taken at the rostro-caudal level at the end of the yolk stalk. Scale bars: 150 µm (i), 10 µm (ii). (E) Lateral views of 28–30 hpf germline mutants (i) and crispants (ii). The urp1∆P;urp2∆P double mutants are maternal zygotic (MZ) mutants. Sibling (sib) and Cas9-only injected embryos served as controls. All embryos were incubated at 28°C, which is a restrictive temperature for cfap298tm304. (F) Quantitative reverse transcriptase PCR (qRT-PCR) analysis of urp1 and urp2 mRNA expression levels in cfap298tm304 and sspob1446 mutants at 28 hpf. n>3 biologically independent samples. Bars represent mean ± s.e.m. Two-tailed student’s t test used to calculate p-values. (G) Schematic of crispant generation and body curve analysis. (H) Quantitation of crispant body curves where bars represent mean ± s.d. for at least three independent clutches and injection mixes. The total number of embryos analyzed is given. *p<0.0001, student’s t test applied. UI – uninjected.
|
|
(A–C) Lateral views of microcomputed tomography reconstitutions of wild-type (A), urp1∆P;urp2∆P (B) and uts2r3b1436 (C) mutants at 3 mpf. (D) Cobb angle measurements for individual fish in the sagittal plane for urp1∆P;urp2∆P and uts2r3b1436 mutants. Circles represent angles for individual curves. (E-E’) Total Cobb angles with each circle representing an individual fish. The mean ± s.d. is shown. (G’) is the data from G parsed for sex. p-Values are given from two-tailed unpaired student’s t tests. (F) The position of curve apex is plotted and shows that most curves are in caudal vertebrae. n=9 and 8 for urp1∆P;urp2∆P and uts2r3b1436 mutants, respectively.
|
|
(A) Lateral views of control fish and age-matched urp1∆P;urp2∆P mutants. Arrows point to forming body curves. (B) Traces of body shape every 2 days for 5 fish per time point from 3 to 17 dpf. (C) Growth curves for control and urp1∆P;urp2∆P mutants were indistinguishable. Arrow shows time of curve onset. Mean ± s.d. is plotted. (D) Microcomputed tomography (µCT) reconstitutions of spines at 1 mpf with heads, fins, and ribs digitally removed. Scale bar: 1 mm. (E) µCT reconstitutions of three pre-caudal and two caudal vertebrae including frontal and lateral views of the highlighted vertebra. No major structural defects such as fusions were observed in urp1∆P;urp2∆P mutants. (F) Vertebral body rostral-caudal length (L1/L2) and dorsal-ventral height (H1/H2) aspect ratios for six vertebrae of wild type (n=4) and urp1∆P;urp2∆P mutants (n=4). Length aspect ratios were significantly more variable in mutants, but height aspect ratios were unchanged (p=0.022 and 0.745, respectively, Bartlett’s test for equal variances). (G) Calcein staining revealed well-structured vertebrae forming in control (standard length 5.7 mm) and urp1∆P;urp2∆P mutant (standard length 5.7 mm) fish. n>30 fish per condition.
|
|
(A–D) Lateral views of microcomputed tomography (µCT) reconstitutions of wild-type (A), cfap298tm304 mutants (B), urp1∆P;urp2∆P double mutants (C), and cfap298tm304;urp1∆P;urp2∆P triple mutants (D). All fish shown are female. Scale bar: 10 mm. (E) Total Cobb angles with each circle representing an individual fish. The mean ± s.d. is shown. p-Values are given from two-tailed unpaired student’s t tests. U — urp1∆P;urp2∆P double mutants; C — cfap298tm304 mutants; UC — cfap298tm304;urp1∆P;urp2∆P triple mutants. (Fi) Dorsal views of µCT reconstitutions with ribs and fins removed. Scale bar: 5 mm. (Fii) Quantitation of degree of lateral curvature for wild type (n=5) and urp1∆P;urp2∆P (n=8), uts2r3b1436 (n=3), cfap298tm304 (n=3), and cfap298tm304;urp1∆Purp2∆P (n=6) mutants. y-axis is the arbitrary units.
|
|
(A–F) Grayscale maximal intensity projection of Sspo-GFP localization in the central canal in 28 hpf embryos (A–C) and 12 dpf adolescents (D–F). RF is denoted by arrow heads in D and F. Arrows point to structures along the central canal that become GFP-positive in cfap298tm304 and uts2r3b1436 mutants. Scale bar: 10 µm. (G) Schematic of temperature shift experiment in which cfap298tm304 mutants are initially raised at permissive temperatures before being shifted to restrictive temperatures at 6 dpf, then imaged at 12 dpf. (H–I) Lateral views of cfap298tm304 (H) and uts2r3b1436 (I) mutants at 12 dpf when Sspo-GFP imaging took place. The white box in H shows the location imaged in D–F.
|