- Title
-
Global mapping of RNA homodimers in living cells
- Authors
- Gabryelska, M.M., Badrock, A.P., Lau, J.Y., O'Keefe, R., Crow, Y.J., Kudla, G.
- Source
- Full text @ Genome Res.
|
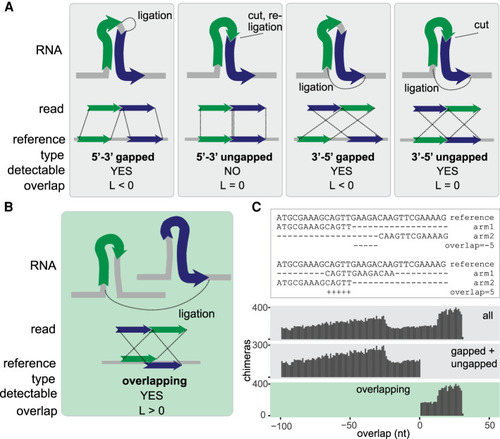
Classification of chimeric reads. (A) Types of nonoverlapping chimeras that may be formed by proximity ligation. The RNA fragments, shown as green and blue arrows, can originate from the same transcript (as shown in the figure) or from distinct transcripts (not shown). The dotted lines in the upper panel indicate ligation sites. (B) Diagram of an overlapping chimera. The RNA fragments originate from two distinct copies of a transcript but are mapped to the same region of the reference gene. (C, top) Examples of nonoverlapping and overlapping chimeric reads mapped to a reference gene. (Bottom) Distribution of the calculated overlap score (L) in all simulated chimeras, simulated nonoverlapping chimeras, and simulated overlapping chimeras. |
|
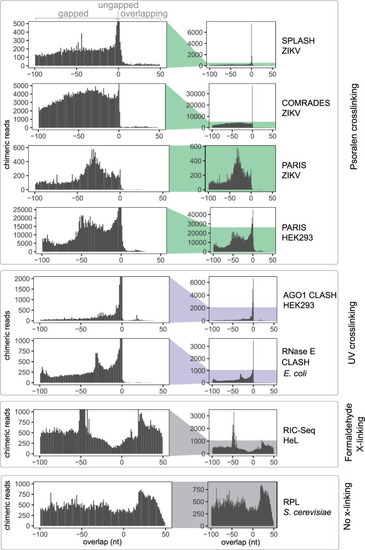
UV and psoralen cross-linking predominantly generate gapped and ungapped chimeras. The distribution of overlap scores (L) across proximity ligation data sets: SPLASH ZIKV (Zika virus) (Huber et al. 2019), COMRADES ZIKV (Ziv et al. 2018), PARIS ZIKV (Li et al. 2018), PARIS HEK293 (human) (Lu et al. 2016), AGO1 CLASH HEK293 (Helwak et al. 2013), RNase E CLASH (Escherichia coli) (Waters et al. 2016), RIC-Seq HeLa (human) (Cai et al. 2020), and RPL (Saccharomyces cerevisiae) (Ramani et al. 2015; Methods). |
|
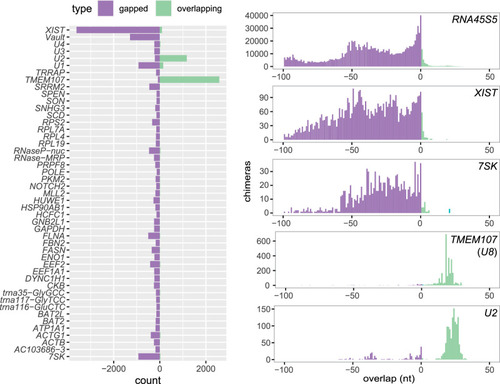
Distribution of overlap scores in individual genes in PARIS experiment. Counts of gapped and ungapped chimeras (purple) and overlapping chimeras (green) in individual genes in PARIS data from human HEK293 cells (Lu et al. 2016) (left), and distribution of the overlap value L in selected genes. |
|
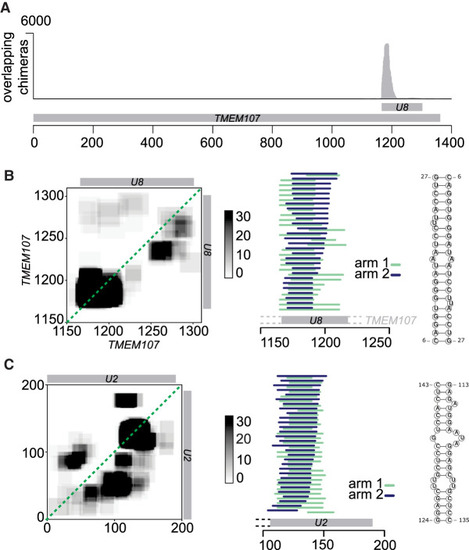
Homodimerization of U8 snoRNA and U2 snRNA. (A) Distribution of overlapping chimeras along the TMEM107 gene in PARIS (HEK293 cells) (Lu et al. 2016). All overlapping chimeras coincide with the position of U8 snoRNA in the 3′ UTR of TMEM107 transcript. (B, left) Coverage map of U8:U8 chimeras. Numbers on the x- and y-axes indicate positions in the TMEM107 transcript. The position of mature U8 snoRNA is indicated by the gray bars. (Middle) Mapping positions of both arms of chimeras along TMEM107. Some chimeras extend beyond the 5′ end of mature U8 (gray bar), indicating that they originate from the U8 precursor molecule. (Right) Predicted base-pairing of U8:U8 homdimer. (C) As above, analysis of U2:U2 chimeras. Coordinates indicate positions along the U2 transcript. |
|
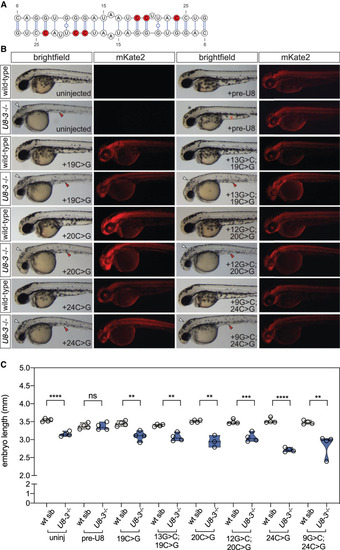
Functional analysis of U8 homodimerization domain. (A) U8:U8 homodimer with positions of selected destabilizing mutations indicated in red. (B) Representative brightfield and fluorescent images of the indicated genotype and exogenous precursor U8 snoRNA variants taken at 48 h post-fertilization. mRNA encoding the mKate2 fluorescent protein was coinjected to show successful uptake of the injected solution into the embryo. White arrowheads denote hydrocephaly, red arrowheads denote aberrant yolk extension, whereas a black asterisk denotes an absence of hydrocephaly and a red asterisk denotes rescued yolk morphology. (C) Quantitation of embryo length for the genotypes and introduced precursor U8 snoRNA variants indicated. (uninj) Uninjected. n = 3–4 embryos per genotype. Black dashed line indicates median value; all data points are shown. |
|
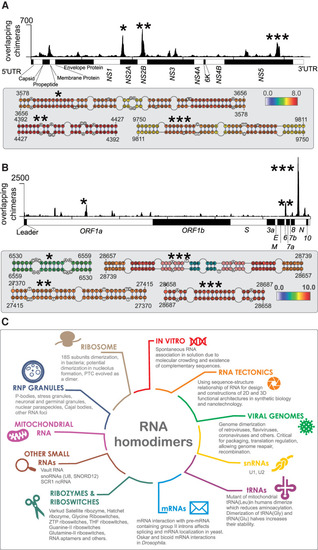
Homodimers of viral genomes. (A, top) Coverage of overlapping chimeras in Zika virus identified by COMRADES (Ziv et al. 2018). The regions with the largest coverage of homodimers are indicated by asterisks. (Bottom) Predicted secondary structures of regions with high coverage of overlapping chimeras. The colors indicate the COMRADES score, that is, the log2 of the number of chimeric reads supporting each base pair. (B) Coverage of overlapping chimeras along the SARS-CoV-2 genome identified by COMRADES (Ziv et al. 2020). The top homodimeric regions and their predicted structures are shown, as described above. (C) Known types of RNA homodimers. |