- Title
-
Long-read sequencing of the zebrafish genome reorganizes genomic architecture
- Authors
- Chernyavskaya, Y., Zhang, X., Liu, J., Blackburn, J.
- Source
- Full text @ BMC Genomics
|
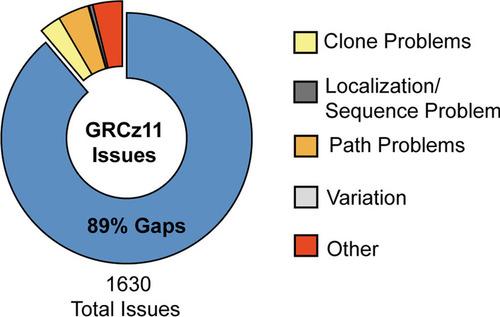
Curated current assembly issues with zebrafish reference genome GRCz11 as reported by the Genome Research Consortium |
|
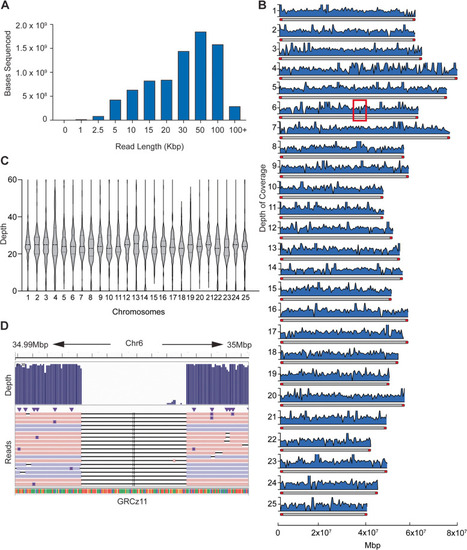
Long-read library run metrics. |
|
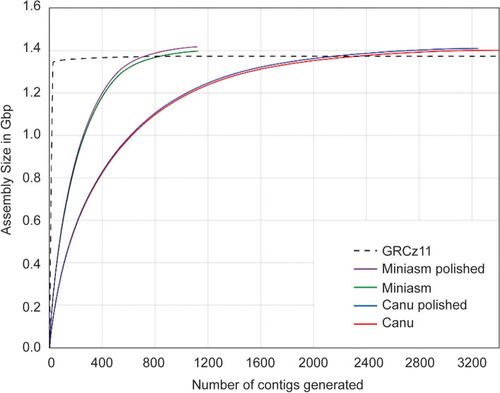
Comparison of total assembly size (Gbp) versus number of contigs generated when using Canu and Miniasm with and without polishing steps. Contigs are ordered largest to smallest, left to right |
|
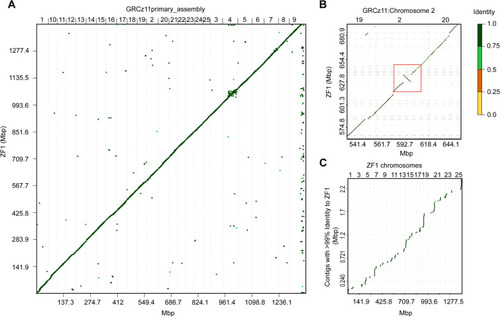
Association plots of similarities and differences between ZF1 assembly and GRCz11 primary assembly. |
|
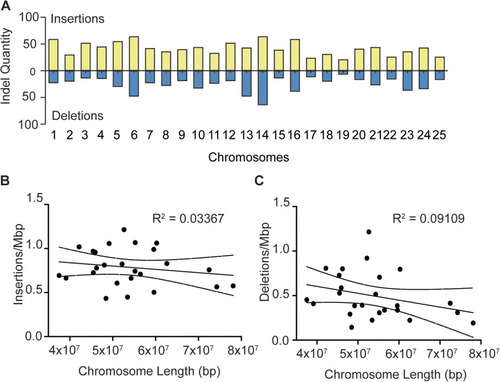
Novel indel distribution in ZF1 assembly. A Frequency of insertions (yellow) and deletions (blue) identified in ZF1 assembly across all chromosomes. B-C Dot plots showing lack of correlation between indel frequency and chromosome length. R value cutoff for correlation was set to 0.6 |
|
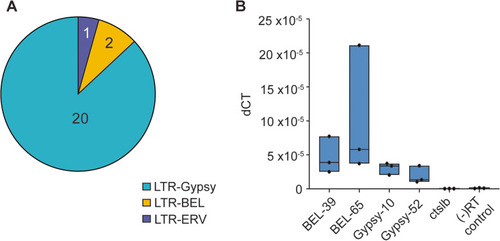
Identification of active retrotransposons in ZF1 assembly. A Results of gene prediction software reveals 23 novel insertions of LTR retrotransposons in the de novo assembly. B Expression by RT-qPCR of select retrotransposons compared to ctslb, which is silenced, and a negative control amplified with primers meant to pick up genomic DNA contamination |