Fig. 2
- ID
- ZDB-FIG-220215-2
- Publication
- Chernyavskaya et al., 2022 - Long-read sequencing of the zebrafish genome reorganizes genomic architecture
- Other Figures
- All Figure Page
- Back to All Figure Page
|
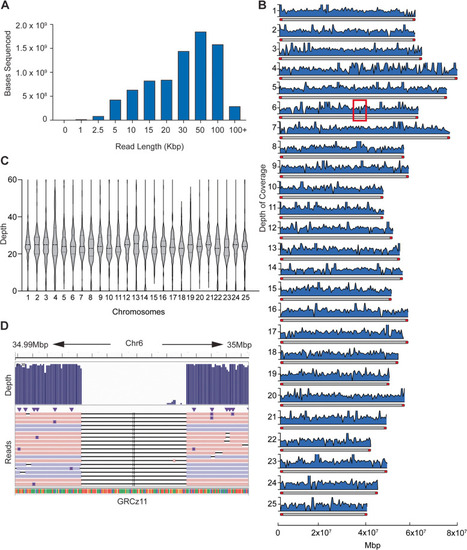
Long-read library run metrics. |