- Title
-
Age-dependent expression changes of circadian system-related genes reveal a potentially conserved link to aging
- Authors
- Barth, E., Srivastava, A., Wengerodt, D., Stojiljkovic, M., Axer, H., Witte, O.W., Kretz, A., Marz, M.
- Source
- Full text @ Aging (Albany NY)
|
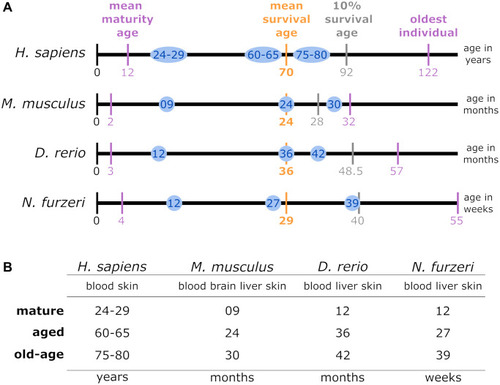
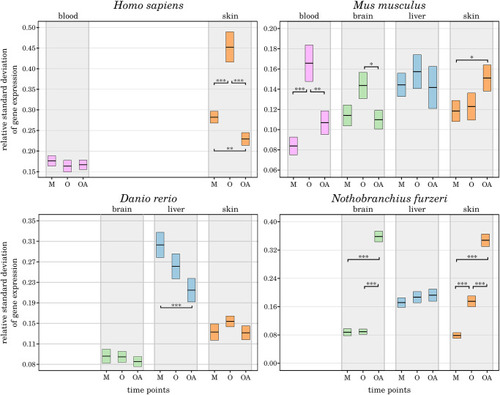
(A) Lifespan comparison: to align the species-specific chronological ages to biological age categories comparable between different species, the total individual’s lifetime, represented by the length of the lifetime axis, was subdivided by index stages. These stages were the biological stages corresponding to maturation, the mean survival age, the 10% survival rate, and the highest age reported for an individual belonging to the respective species. The time intervals between the resulting intersections were normalized linearly for the mean survival age. Thus, the sampling time points examined in this study for a specific species (blue circles) matched the biological age category in all the other species. (B) Data sampling scheme and categorization of the high-throughput transcriptomic data according to age parameters: for each of the 4 species of interest, up to 4 tissue types were sampled at mature, aged and old-age stages, from which 3 comparisons were deduced to identify DEGs during early aging (mature vs. aged), late aging (mature vs. old-age), and longevity (aged vs. old-age). |
|
|
|
|
|
|
|
|
|
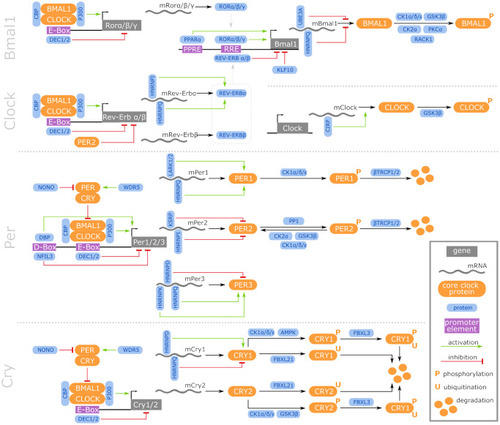
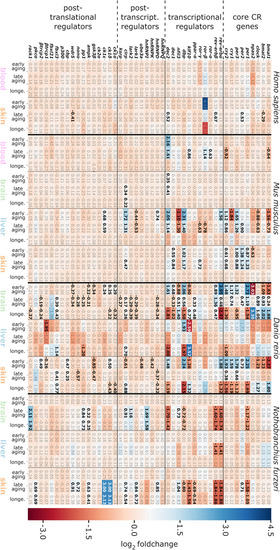
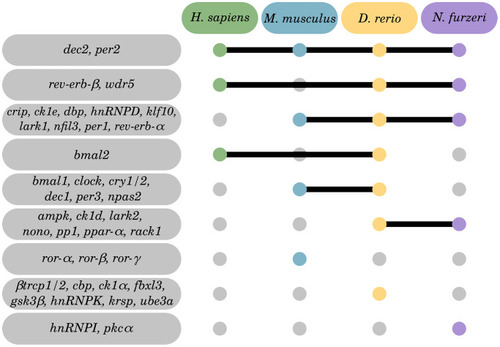
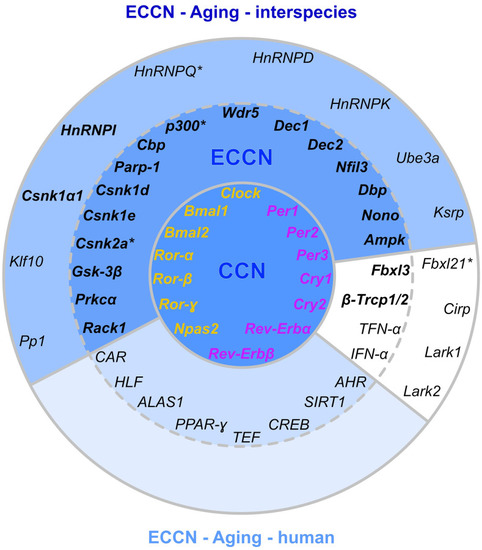
Matching of our CR-related DEGs evidenced to be regulated with aging with a curated human CR network. The core clock network (CCN; inner ring) is comprised of a positive (orange) and negative (purple) branch of core clock regulators and has been expanded to an extended core clock network (ECCN; middle ring) as determined by chronobiological meta-analyses in humans [84]. The majority of CR-related genes identified in the present study overlaps (represented in bold) with the human CCN (100% overlap) and ECCN (65.5% overlap). The ECCN was complemented by 13 CR-related genes defined by our aging-related interspecies approach (outer ring), two of which overlapped with newly designated CR genes (bold in outer ring [84]). By mapping to a curated biomedical aging database [101], 85% of the presented interspecies (dark blue background) or human (light blue background) CR-related genes were identified to be aging-associated. The remaining factors (white background) have not yet been annotated to aging but might represent new candidates. *, curated CR-related genes without age-related differential expression within the interspecies comparison. |