- Title
-
The localization of amyloid precursor protein to ependymal cilia in vertebrates and its role in ciliogenesis and brain development in zebrafish
- Authors
- Chebli, J., Rahmati, M., Lashley, T., Edeman, B., Oldfors, A., Zetterberg, H., Abramsson, A.
- Source
- Full text @ Sci. Rep.
|
Expression pattern of EXPRESSION / LABELING:
|
|
Localization of App protein to cilia of the olfactory sensory neurons and otic vesicle in 31 hpf larvae. Cilia as shown by immunostaining for acetylated tubulin (magenta) and App (green) of the olfactory sensory neurons in the nose epithelium ( EXPRESSION / LABELING:
|
|
App localizes to the cilia decorating the ventricle of larvae and ependymal cells in adult zebrafish. (A) Schematic representations of head and ventricle morphology in 30 hpf zebrafish larvae, dorsal view. (B) Dorsal view of ventricle immunostained for App (green) and acetylated tubulin (magenta) in 30 hpf WT zebrafish larvae. (C) Close-up of cilia (magenta) and App (green). (D) Negative controls for immunofluorescence with secondary antibodies without anti-acetylated tubulin and anti-App primary antibodies. Cell nuclei stained with DAPI (blue). (E) Schematic outline of adult zebrafish brain, lateral view. (F–G) Coronal section of adult zebrafish brain and the central canal (see asterisk). Cell nuclei labeled with DAPI (blue), acetylated tubulin (magenta), App (green). (G) Ependymal motile cilia (magenta) of the central canal with App (green) accumulation along cilia. Magnification: (B,F) = 10×, (D) = 40×, (C,G) = 60×. Scale bar: (B,D) = 50 µm, (C) = 1 µm, (F) = 500 µm, (G) = 10 µm. |
|
APP is localized to the ependymal cilia in adult mouse. ( |
|
APP is localized to human ependymal cilia. ( |
|
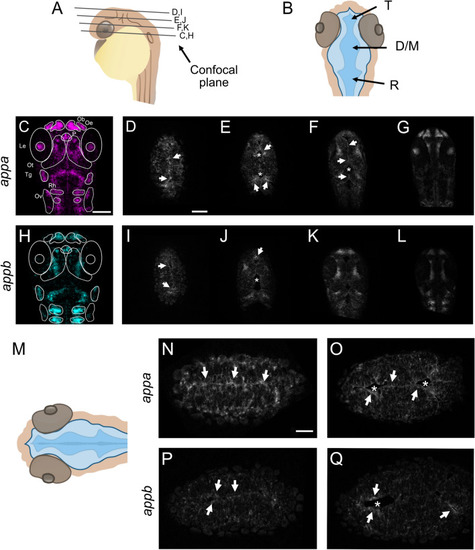
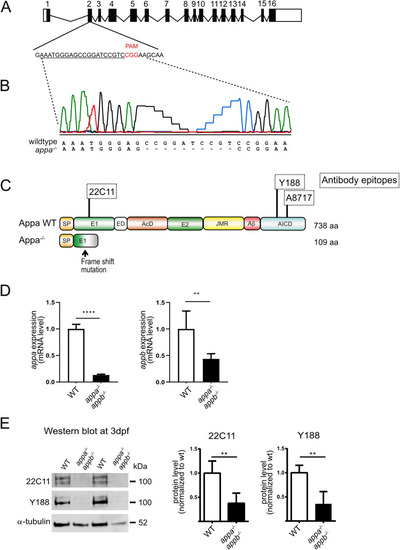
Generation of appa EXPRESSION / LABELING:
PHENOTYPE:
|
|
Longer cilia of dorsal brain ventricle neuroepithelium in PHENOTYPE:
|
|
Structural integrity of ependymal cilia in WT and PHENOTYPE:
|
|
The taken from brain ventricles of dextran injected 2 dpf zebrafish larvae ( PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|