- Title
-
Probing Synergistic Targets by Natural Compounds for Hepatocellular Carcinoma
- Authors
- Gao, J., Yin, Z., Wu, Z., Sheng, Z., Ma, C., Chen, R., Zhang, X., Tang, K., Fei, J., Cao, Z.
- Source
- Full text @ Front Cell Dev Biol
|
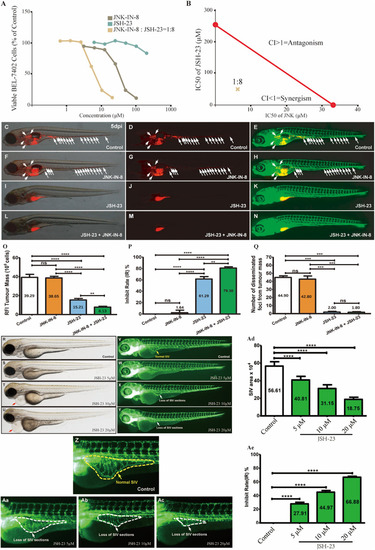
CBE synergisti cally inhibited the growth of BEL-7402 cells. |
|
The detection of key players in CBE treatment based on differential signaling network. |
|
Coinhibition of NF-κB and c-JUN synergistically suppressed HCC growth and |
|
Liver cancer patients with low NF-κB and JNK expression showed a better prognosis and decreased proliferation. |
|
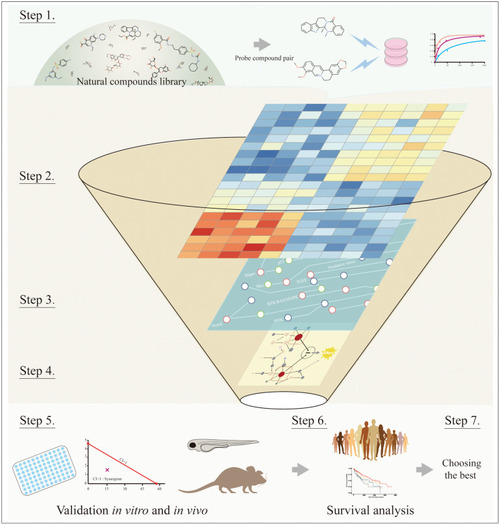
The workflow of systematic processes to identify key synergy-mediated players. Step 1: Choosing the natural compound pair from the natural compounds library. Step 2: Obtaining DEGs from transcriptomics or proteomics profiling. Step 3: Mapping DEGs to cancer signaling transduction networks. Step 4: Inferring key synergy-mediating targets among DEGs based on network topology and empirical experience. Step 5: Validating suspected targets through |