- Title
-
A stochastic oscillator model simulates the entrainment of vertebrate cellular clocks by light
- Authors
- Kumpošt, V., Vallone, D., Gondi, S.B., Foulkes, N.S., Mikut, R., Hilbert, L.
- Source
- Full text @ Sci. Rep.
|
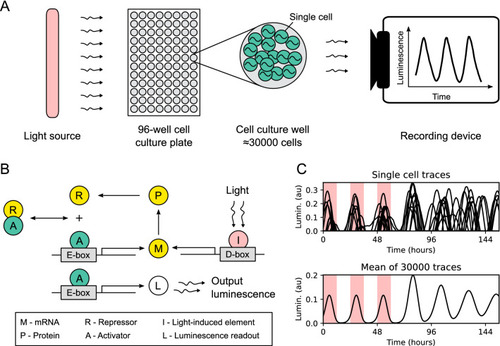
Experiment setup and simulation of core circadian clock dynamics in zebrafish cell cultures. ( |
|
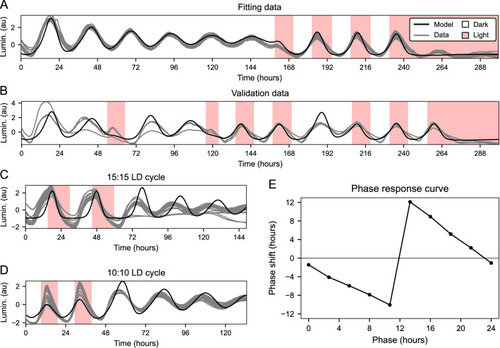
Simulation of |
|
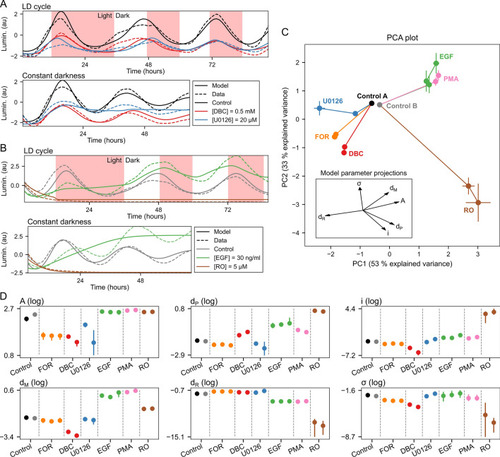
Characterization of compound effects by refitting of model parameters. ( |