- Title
-
Automatic Segmentation and Cardiac Mechanics Analysis of Evolving Zebrafish Using Deep Learning
- Authors
- Zhang, B., Pas, K.E., Ijaseun, T., Cao, H., Fei, P., Lee, J.
- Source
- Full text @ Front Cardiovasc Med
|
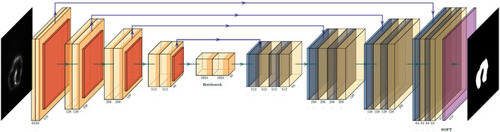
U-net convolution neural network (CNN) architecture utilized to generate the binary mask of the intracardiac domain of zebrafish. Each box represents a multichannel feature map that allows for efficient and accurate extraction of anatomical features. In our specific application, the input was a 512 × 512 pixel map. |
|
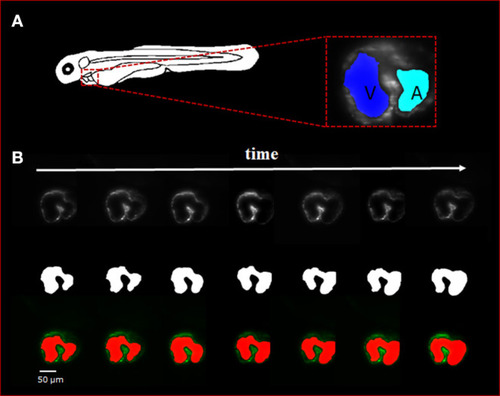
Sequence of selected light-sheet fluorescent microscopy (LSFM) images with the manual hand segmentation mask from 4 dpf zebrafish heart. |
|
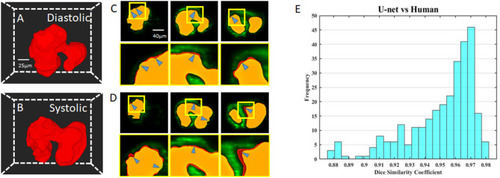
Comparison of U-net based autosegmentation to manual hand segmentation. |
|
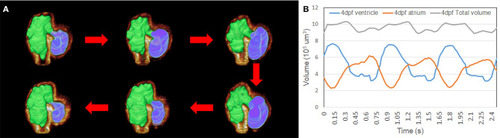
Subdivision of atrium and ventricle. |
|
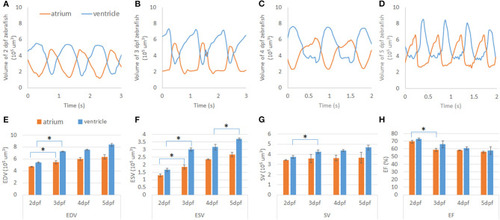
Representation of the contraction to dilation of zebrafish heart volume change over time. |
|
Cardiac mechanics analysis of developing zebrafish heart. |