- Title
-
Myomixer is expressed during embryonic and post-larval hyperplasia, muscle regeneration and differentiation of myoblats in rainbow trout (Oncorhynchus mykiss)
- Authors
- Perello-Amoros, M., Rallière, C., Gutiérrez, J., Gabillard, J.C.
- Source
- Full text @ Gene
|
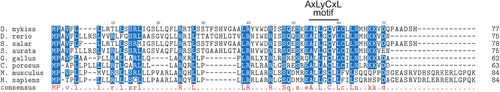
The sequence alignment of vertebrate Myomixer proteins. The alignment was performed from the complete protein sequences using ClustalW multiple alignment tool. The amino acid residues present in all sequences are uppercase and lowercase when present in at least 6 sequences. The AxLyCxL motif was indicated: x corresponds to leucine, isoleucine, valine and y denotes serine, threonine, alanine or glycine. Accession numbers are as follows: O. mykiss, QII57370; D. rerio, P0DP88 ; S. salar, XM-014180492; S. aurata, ERR12611_isotig14560 (http://sea.ccmar.ualg.pt:4567/); G. gallus, CD218366.1; C. porosus, XP_019405207; M. musculus, Q2Q5T5 and H. sapiens, A0A1B0GTQ4. |
|
Phylogenetic analysis of Myomixer in tetrapods and teleosts. The phylogenetic tree was constructed from a multiple alignment of the complete sequences of the proteins using the neighbour-joining method. The numbers at the tree nodes represent percentage of bootstrap values after 500 replicates. |
|
Patterns of myomixer expression during embryonic development. (A-F) Embryos were analyzed by whole mounted in situ hybridization at day 10, 14 and 18 post fertilization. The corresponding vibratome section (35 µm) was presented for each stage. Asterisks indicate the dorsal and ventral domains of the myotome and arrowhead indicates the dermomyotome-like epithelium. (G-I) Double in situ hybridization for pax7 and myomixer of 17 dpf embryo sections. The nuclei are counter-stained with DAPI and arrowhead indicates the dermomyotome-like epithelium. (J-L) The expression of myomixer and myomaker in muscle of 1 g and 20 g trout was also studied using in situ hybridization on cross sections (7 µm). |
|
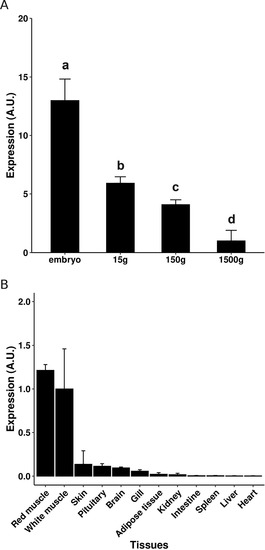
Expression of myomixer in tissues and white muscle of different-weight trout. The quantification of myomixer expression was performed by qPCR analysis in muscle of different weight trout (A) and in several tissues (150 g; B). The qPCR results are presented as a ratio of myomixer and eF1a expression, and the bars represent the standard error. The letters (a-d) in A indicate the significant differences between means (p < 0.05; Kruskal–Wallis rank test followed by the Wilcoxon-Mann-Whitney test). |
|
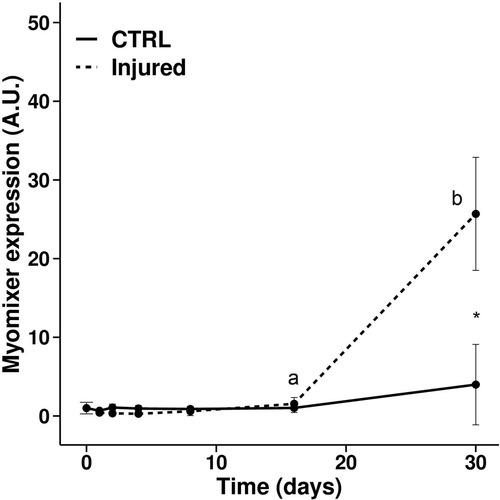
Expression of myomixer during muscle regeneration in trout. Gene expression profile of myomixer during muscle regeneration in rainbow trout normalized with eF1a expression. Bars represent the standard error and the letters indicate the significant differences between means within the same treatment (control or injured). The asterisk indicates significant differences between treatments at a given time. Statistical significance (p < 0.05) was determined using the Kruskal–Wallis rank test followed by the Wilcoxon-Mann-Whitney test. |
|
Expression of myomixer and myomaker during trout satellite cell differentiation. The cells were cultivated in proliferative medium (PM) and then in differentiation medium for 1, 2, and 3 days (DM1, DM2, and DM3). The qPCR results are normalized with eF1a expression and bars represent the standard error. Different letters indicate significant differences between means. Statistical significance (p < 0.05) was determined using the Kruskal–Wallis rank test followed by the Wilcoxon-Mann-Whitney test. |
Reprinted from Gene, 790, Perello-Amoros, M., Rallière, C., Gutiérrez, J., Gabillard, J.C., Myomixer is expressed during embryonic and post-larval hyperplasia, muscle regeneration and differentiation of myoblats in rainbow trout (Oncorhynchus mykiss), 145688, Copyright (2021) with permission from Elsevier. Full text @ Gene