- Title
-
son is necessary for proper vertebrate blood development
- Authors
- Belmonte, R.L., Engbretson, I.L., Kim, J.H., Cajias, I., Ahn, E.E., Stachura, D.L.
- Source
- Full text @ PLoS One
|
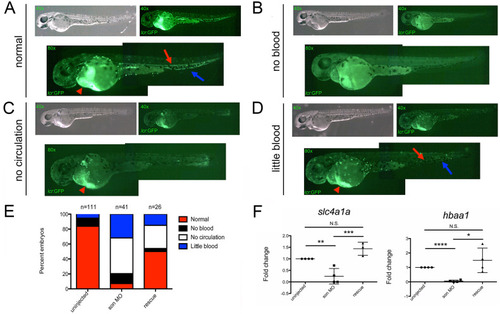
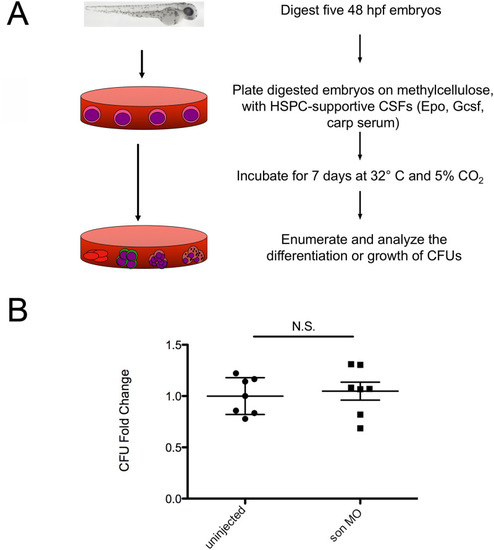
Representative images of 48hpf |
|
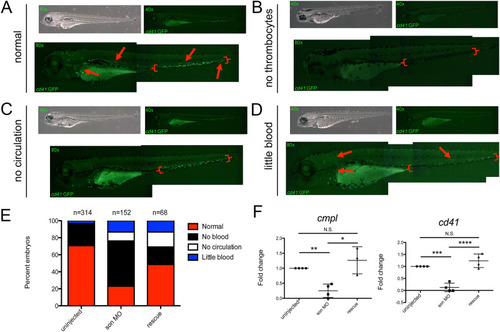
Representative images of 72hpf cd41:GFP embryos injected with son MO at the one-cell-stage of development show four common phenotypic categories associated with MO injection. Category titles are listed to the left of each image. Brightfield images are shown in top left (40x), and fluorescent images are shown in top right (40x). Zoomed in fluorescent images are shown below (80x); each green dot in the animal is a labelled thrombocyte. Red arrows indicate fluorescent “trails” made by moving thrombocytes flowing though the vasculature. Red brackets indicate the caudal hematopoietic region where the majority of thrombocytes reside at this time. (A) Representative images of normal thrombocyte numbers and circulating thrombocytes. (B) Representative animals with no thrombocytes, (C) no thrombocytes circulating in the animal (no circulation), and (D) reduced numbers of thrombocytes, some of which are circulating (little blood). (E) Quantitation of phenotypes shown. Number of embryos analyzed is shown above the chart. (F) qRT-PCR for cmpl (left) and cd41 (right) was performed. Each point represents ten embryos randomly selected from uninjected, MO-injected (son MO), or MO-injected with son RNA (rescue) conditions that were analyzed by qRT-PCR. Middle lines represent mean and error bars represent SD. * represents p = 0.04, ** represents p = 0.006, *** represents p = 0.002, **** represents p = 0.001, N.S. represents no significance. |
|
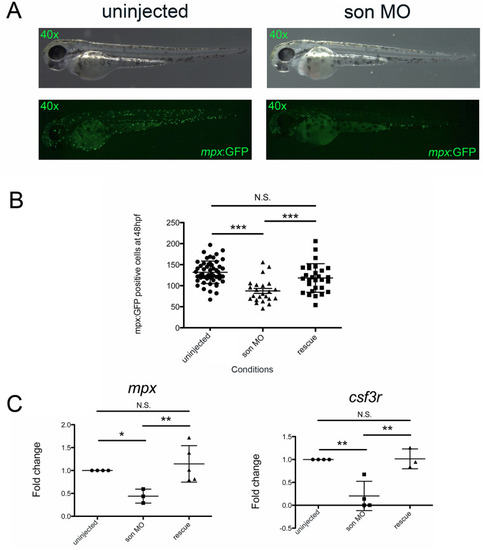
( |
|
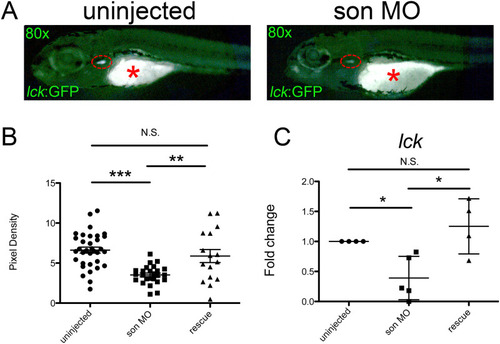
( |
|
( |