- Title
-
A conserved acetylation switch enables pharmacological control of tubby-like protein stability
- Authors
- Kerek, E.M., Yoon, K.H., Luo, S.Y., Chen, J., Valencia, R., Julien, O., Waskiewicz, A.J., Hubbard, B.P.
- Source
- Full text @ J. Biol. Chem.
|
|
|
|
|
|
|
|
|
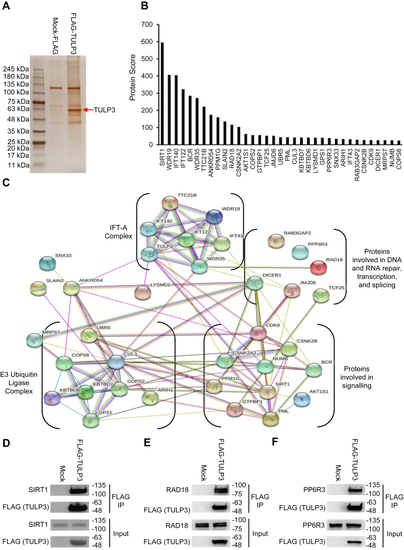
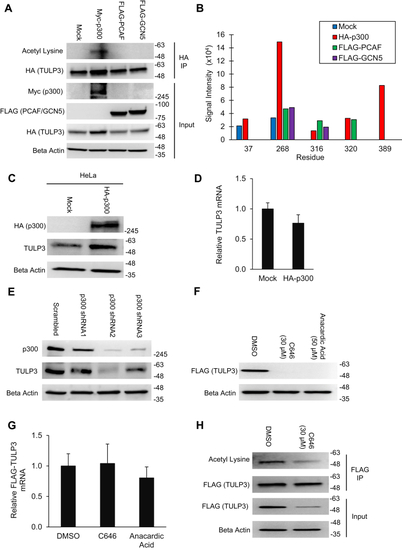
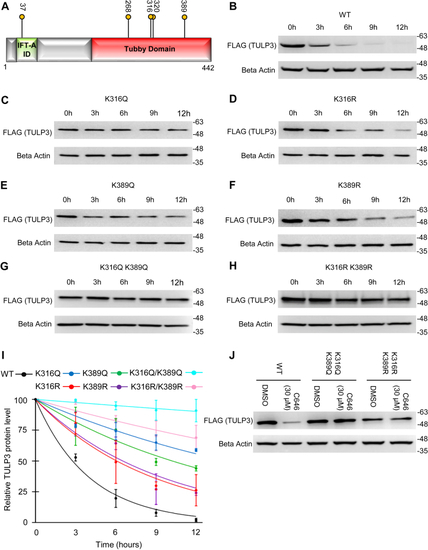
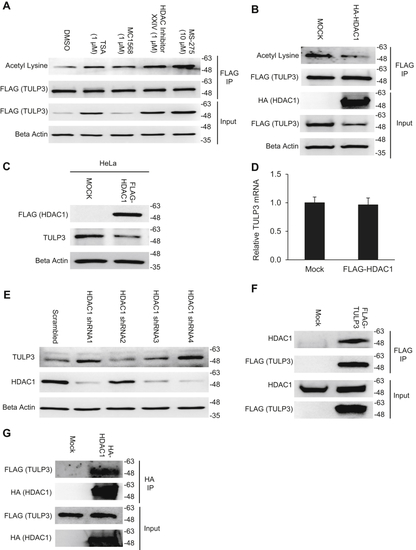
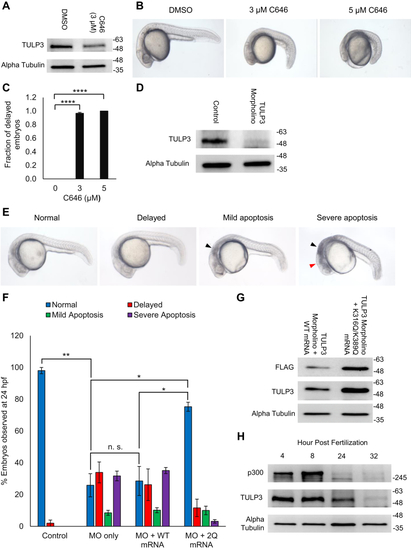
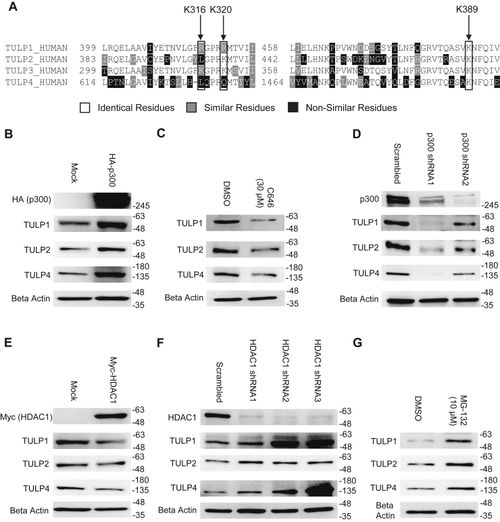
TULP3 protein stability is regulated by polyubiquitination and by Cullin-3. A, western blot showing TULP3 levels in HeLa cells treated with 10 μM MG-132 for the time intervals indicated. B, TULP3 protein levels in the absence or presence of 10 μM MG-132 and transfected with a plasmid encoding HA-p300 as indicated. Cells were harvested 48 h posttransfection following 9 h of MG-132 treatment. C, immunoblot analysis of ubiquitination levels on FLAG-TULP3 immunoprecipitated from 293T cells treated with MG-132 and transfected with plasmids encoding HA-Ubiquitin or Myc-p300 as indicated. Cells were harvested 48 h posttransfection following 9 h of MG-132 treatment. D, western blot showing ubiquitination levels on FLAG-TULP3 immunoprecipitated from 293T cells treated with MG-132 and transfected with plasmids encoding HA-Ubiquitin or Myc-HDAC1 as indicated. Cells were harvested 48 h posttransfection following 9 h of MG-132 treatment. E, schematic outlining key functional domains in TULP3 and the location of ubiquitination sites identified by mass spectrometry. IFT-A ID denotes Intraflagellar Transport Complex A interacting domain. F, western blot showing coimmunoprecipitation of FLAG-TULP3 with Cullin-3 in 293T cells. G, immunoblot showing levels of FLAG-TULP3 protein in 293T cells in the absence or presence of C646 following transfection with a pool of siRNAs targeting Cullin-3 or a scrambled control. Cells were collected 72 h posttransfection; 30 μM C646 was added to cells 48 h posttransfection as indicated. |
|
|
|
|