- Title
-
PRL3-DDX21 Transcriptional Control of Endolysosomal Genes Restricts Melanocyte Stem Cell Differentiation
- Authors
- Johansson, J.A., Marie, K.L., Lu, Y., Brombin, A., Santoriello, C., Zeng, Z., Zich, J., Gautier, P., von Kriegsheim, A., Brunsdon, H., Wheeler, A.P., Dreger, M., Houston, D.R., Dooley, C.M., Sims, A.H., Busch-Nentwich, E.M., Zon, L.I., Illingworth, R.S., Patton, E.E.
- Source
- Full text @ Dev. Cell

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
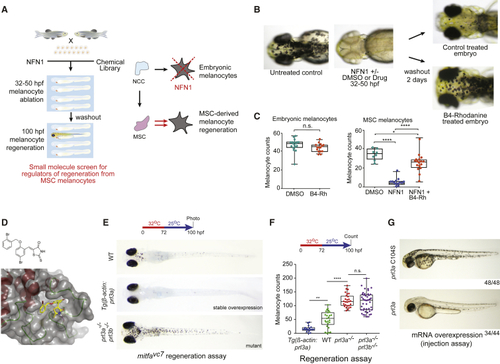
Prl3a Is an Inhibitor of Melanocyte Regeneration in Zebrafish (A) Schematic of a small-molecule screen for regulators of MSC-derived melanocytes in zebrafish. MSC, melanocyte stem cell; NCC, neural crest cells. (B) Images of zebrafish embryos treated with NFN1 ± DMSO or B4-Rh and after washout. (C) Quantification of zebrafish melanocytes during normal development (n.s., not significant, Student’s t test) or in a NFN1-regeneration assay (ANOVA using Tukey’s analysis; ∗p value = 0.0131; ∗∗∗∗p ≤ 0.0001). (D) Predicted binding of B4-Rh (yellow sticks) in the NMR model of PRL3 (gray transparent surface and secondary structure; red, helix; green, loop). Purple-dashed line: predicted hydrogen bond to E50. All other protein-ligand interactions are apolar. The ligand sits in a hydrophobic pocket formed by residues: V48, C49, W68, P69, A74, P75, P77, V80, A111, V113, and the methylene groups of the side chain of Q145. These residues, and E50, are conserved in zebrafish Prl3a. (E and F) (E) Images and (F) quantification of wild type, (G) RNA overexpression of See also |
|
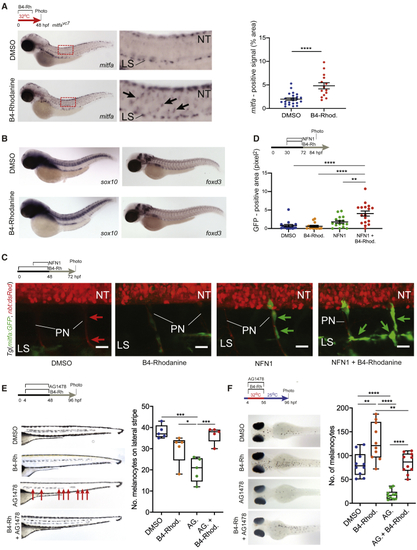
Inhibition of Prl3 Leads to Premature Melanoblast Expansion during Regeneration (A) (B) (C) Confocal imaging of the MSC lineage during treatments specified. (D) Quantification of area of GFP expression (pixels2) on individual peripheral nerves of 84 hpf (E) Lateral stripe melanocytes following specified treatments. AG.: AG1478. B4-Rhod: B4-Rhodanine. Red arrows: missing melanocytes; faint melanocytes from opposite side of the body are also visible. Quantification of melanocytes on the lateral stripe of embryos at 96 hpf, ∗p = 0.0364; ∗∗∗p = 0.0003 and 0.0006 for DMSO versus AG. and AG. versus AG. + B4-Rhod, respectively, ANOVA using Tukey’s analysis. (F) Melanocyte regeneration in |
|
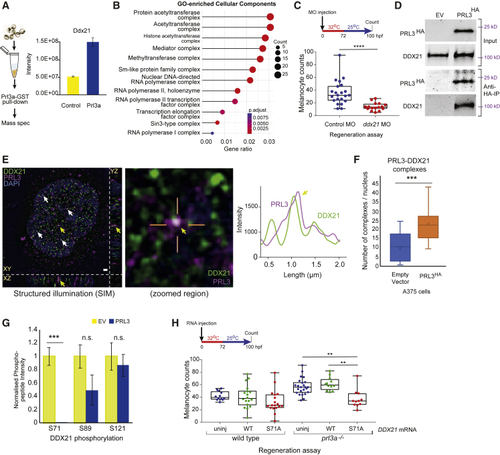
Prl3a Interacts with Ddx21 in Zebrafish and Melanoma Cell Nuclei (A) Experimental overview and intensity quantification of Ddx21 peptides from Prl3a-GST pull-downs. Bars and error bars are mean ± SEM. See also (B) Cellular component GO-enrichment analysis of Prl3a-interacting proteins identified by co-immunoprecipitation and mass-spectrometry (adjusted p values: Benjamini-Hochberg test). (C) Quantification of melanocytes in a (D) Co-immunoprecipitation of human HA-tagged PRL3 protein from A375 cells: empty vector (EV) and PRL3-expressing stable transfected cells. (E) Structured illumination microscopy (SIM) of endogenous PRL3 (magenta) and DDX21 (green) in C092 melanoma cells. Scale bar, 1 μm. Co-localization indicated (arrows); zoomed image shows PRL3-DDX21 co-localization with line scan, and intensity plot profile of line scan showing signal overlap (yellow arrow). DAPI: blue. See also (F) Quantification of PRL3-DDX21 complexes per nucleus identified by SIM in A375 melanoma cells expressing empty vector or HA-tagged PRL3 (∗∗∗p < 0.001, error bars: x indicates the mean; Student's t test). See also (G) Mass spectrometry of DDX21 phosphorylation sites in EV control cells (n = 5 samples) and cells expressing PRL3 (n = 7 samples) (∗∗∗p < 0.001; n.s., not significant; Bars and error bars are mean ± SEM; Student's t test). See also (H) Quantification of regenerating melanocytes in wild-type or |
|
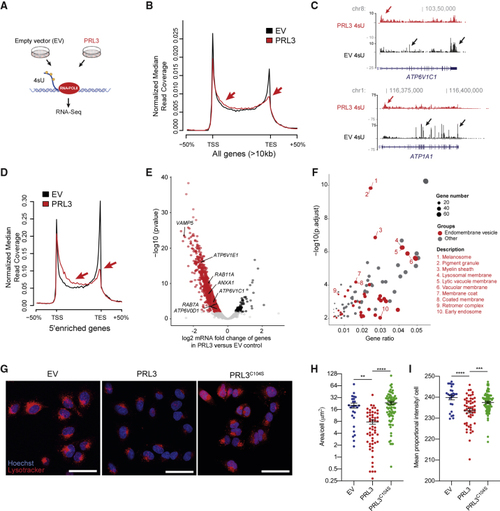
PRL3 Restrains Transcriptional Elongation of Endomembrane Vesicle Genes (A) Schematic of 4sU protocol to detect nascent RNA transcripts. (B) 4sU transcriptional profile for long genes in EV (empty vector) versus (C) 4sU RNA-seq transcript coverage for (D) 4sU transcriptional profile for 5′-enriched genes in control and (E) Volcano plot of differentially expressed genes in PRL3 versus EV control A375 cells by DESeq2-analysis using 4sU nascent RNA-seq data. See also (F) Bubble plot of GO cellular components in the (G) A375 melanoma cells expressing EV, PRL3, and PRL3 (C104S) stained with LysoTracker (red) and hoechst (blue). Scale bars: 50 μm. (H) Quantification of the area of LysoTracker-positive vesicles per cell. ANOVA using Tukey’s analysis, ∗∗p = 0.0015; ∗∗∗∗p < 0.0001. Line and error bars, mean ± SEM. (I) Quantification of the proportional mean intensity of LysoTracker-positive vesicles per cell. ANOVA using Tukey’s analysis, ∗∗∗p = 0.0005; ∗∗∗∗p < 0.0001. Line and error bars, mean ± SEM. See also |
|
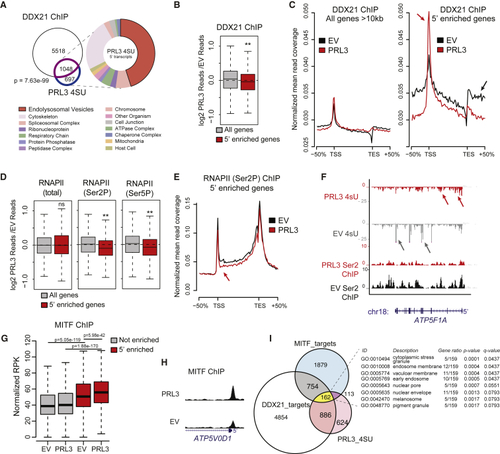
PRL3 Restrains Pol II Transcriptional Elongation of DDX21 Target Genes at MITF Targets (A) Venn diagram of overlapping genes between DDX21 ChIP targets (purple) and PRL3 5′-enriched genes (blue) (p = 7.63 e-99, Fisher's exact test). GO enrichment analysis of cellular components (false discovery rate [FDR] < 0.05). See also (B) DDX21 ChIP-seq data as log2 ratios of PRL3 over-expressing (PRL3) versus empty vector (EV) control for all long (All genes) and 5'-enriched genes., ∗∗p = 1.61 × 10−8, Wilcoxon signed-rank test. (C) DDX21 ChIP-seq read profile for all long or 5′-enriched genes in control EV and PRL3-expressing cells. In 5′-enriched genes, PRL3 causes a net accumulation of DDX21 to the 5′ end (red arrows) versus EV control-treated cells (black arrow). (D) Boxplots of RNAP II (total, Ser2P, and Ser5P) ChIP-seq data shown as ratios of PRL3 versus control read depth for all long genes and 5′-enriched genes. (5′-enriched p = 0.83, 1.26 × 10−18 3.93 × 10−11 for total, Ser2P, and Ser5P RNAP, respectively; Wilcoxon signed-rank test). (E) RNAP II (Ser2P) ChIP-seq profile for 5′-enriched genes in EV control and PRL3-expressing cells. RNAP II (Ser2P) signal is decreased at the 5′ end (red arrow) and throughout the gene body versus control cells (black). (F) 4sU RNA-seq and RNA PolII (Ser2P) ChIP-seq transcript coverage for (G) MITF ChIP-seq signal surrounding the TSS (±1 kb) for non-5′-enriched (gray) and 5′-enriched (red) genes (p values determined using paired and un-paired Wilcoxon rank sum tests for within and between gene set comparisons, respectively). (H) MITF ChIP-seq profile of (I) Venn diagram of DDX21 ChIP targets (white, 6,566 total), PRL3 5′-accumulated genes (pink, 1,745 total), and MITF ChIP targets (blue, 2,908 total). 162 genes overlap the three groups, highlighted in yellow (p = 6.67e−06, hypergeometric test). Table lists cellular component GO enrichment analysis of these 162 genes (FDR < 0.05). See also |
|
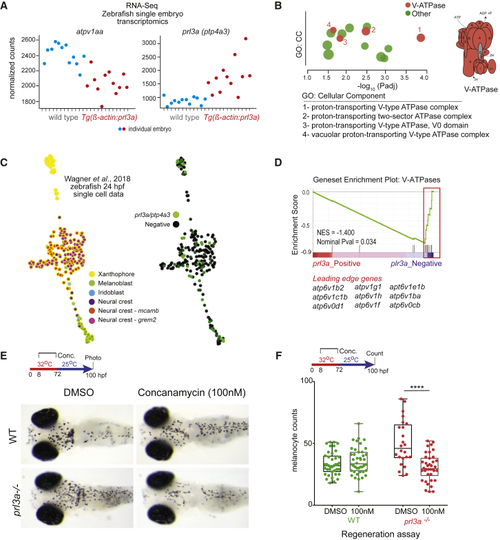
(A) Single embryo transcriptomics of heterozygote (B) g:Profiler output shows V-ATPase complex GO enrichment for (C) K-nearest neighbor graph (SPRING webtool) of (D) GSEA plotting enrichment of (E and F) (E) Images and (F) quantification of melanocytes in wild-type (WT) or |
|
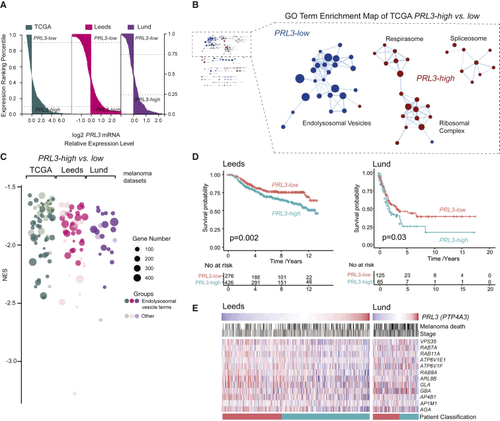
A (A) Patient samples ranked by (B) GO enrichment analysis (cellular compartment) of (C) Bubble plot of GO cellular components in (D) Kaplan Meier survival curves of (E) Heatmap of endolysosomal genes ranked by See also |
Reprinted from Developmental Cell, 54(3), Johansson, J.A., Marie, K.L., Lu, Y., Brombin, A., Santoriello, C., Zeng, Z., Zich, J., Gautier, P., von Kriegsheim, A., Brunsdon, H., Wheeler, A.P., Dreger, M., Houston, D.R., Dooley, C.M., Sims, A.H., Busch-Nentwich, E.M., Zon, L.I., Illingworth, R.S., Patton, E.E., PRL3-DDX21 Transcriptional Control of Endolysosomal Genes Restricts Melanocyte Stem Cell Differentiation, 317-332.e9, Copyright (2020) with permission from Elsevier. Full text @ Dev. Cell