- Title
-
A chiral selectivity relaxed paralog of DTD for proofreading tRNA mischarging in Animalia
- Authors
- Kuncha, S.K., Mazeed, M., Singh, R., Kattula, B., Routh, S.B., Sankaranarayanan, R.
- Source
- Full text @ Nat. Commun.
|
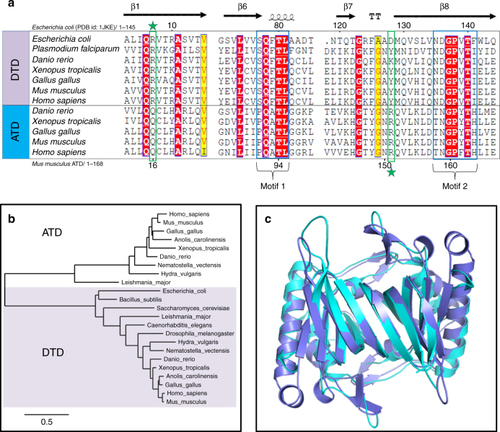
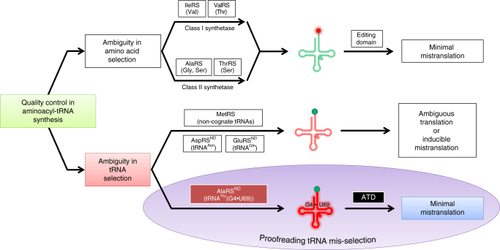
ATD is a variant of DTD. |
|
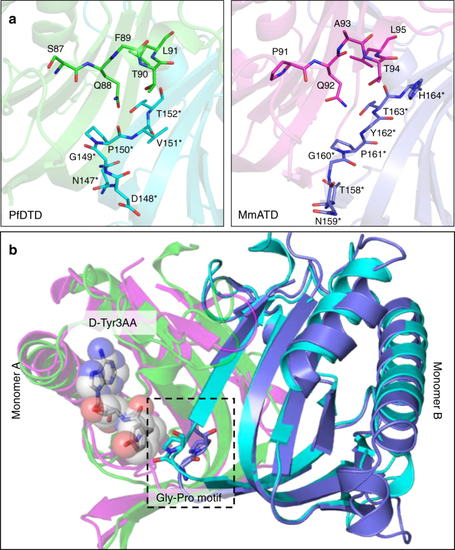
ATD has similar active site features as compared to DTD’s. |
|
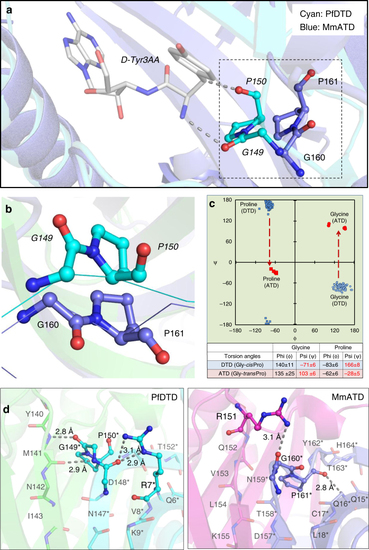
ATD has a Gly- |
|
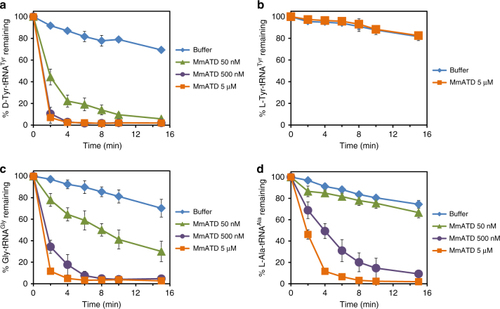
ATD displays relaxation of substrate chiral specificity. |
|
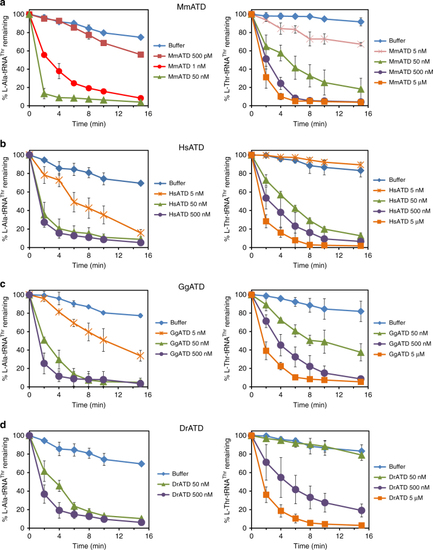
Proofreading of L-Ala-tRNAThr(G4•U69) by ATD is conserved across organisms. Deacylation of |
|
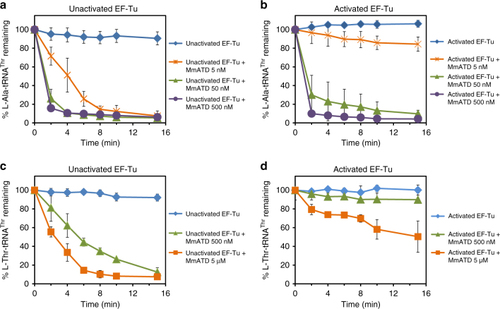
EF-Tu confers protection on L-Thr-tRNAThr(G4•U69) against ATD. Deacylation of |
|
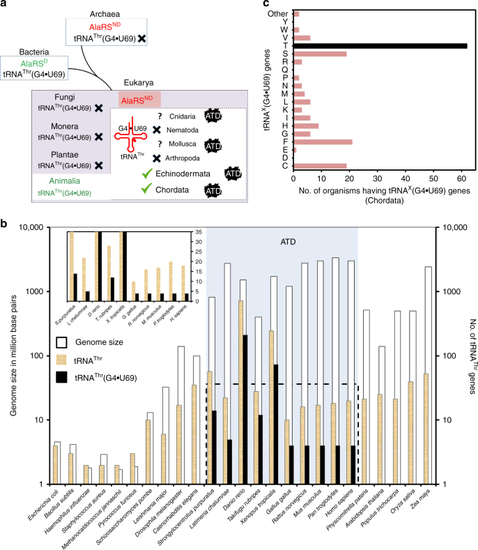
Enrichment of tRNAThr(G4•U69) genes and presence of ATD show strict association. |
|
ATD is a unique and dedicated proofreading factor that rectifies a critical tRNA selection error. Model for mis-selection and consequent misacylation of tRNAThr(G4•U69) with |