- Title
-
Cytokinetic bridge triggers de novo lumen formation in vivo
- Authors
- Rathbun, L.I., Colicino, E.G., Manikas, J., O'Connell, J., Krishnan, N., Reilly, N.S., Coyne, S., Erdemci-Tandogan, G., Garrastegui, A., Freshour, J., Santra, P., Manning, M.L., Amack, J.D., Hehnly, H.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
|
|
|
|
|
|
|
|
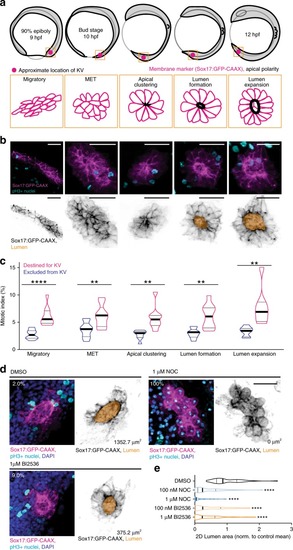
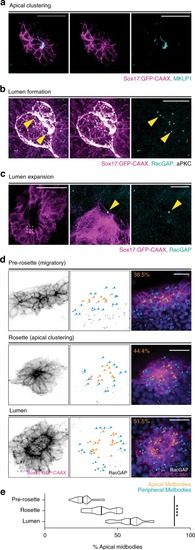
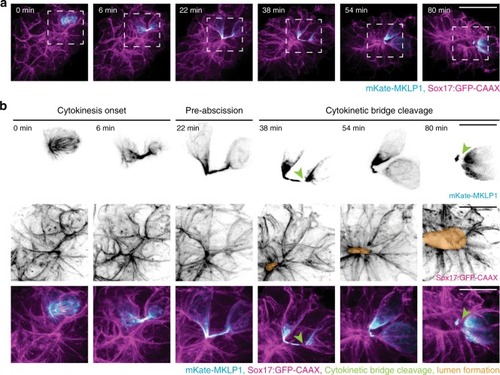
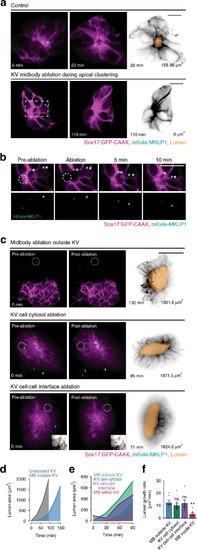
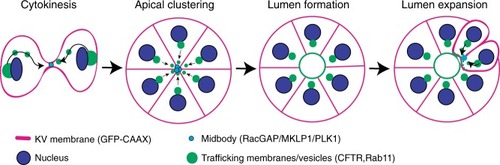
Model depicting lumen formation through Rab11-mediated vesicle transport to the cytokinetic bridge. KV membrane (GFP-CAAX—magenta), midbodies (RacGAP/MKLP1/PLK1—cyan), vesicles (CFTR/Rab11—green), and nuclei (blue) are shown. |