- Title
-
Evolutionary and Molecular Characterization of liver-enriched gene 1
- Authors
- Dang, Y., Wang, J.Y., Liu, C., Zhang, K., Jinrong, P., He, J.
- Source
- Full text @ Sci. Rep.
|
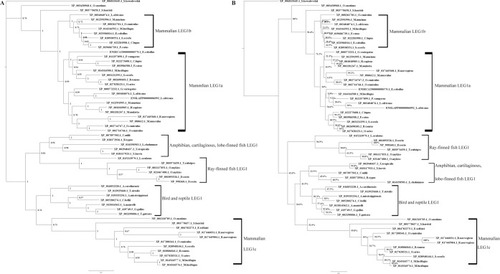
Phylogenetic analysis of LEG1 protein sequences in representative vertebrates performed using Bayesian ( |
|
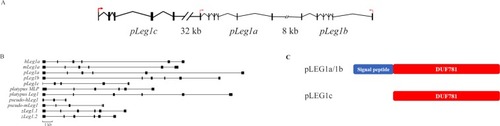
The genomic organization of |
|
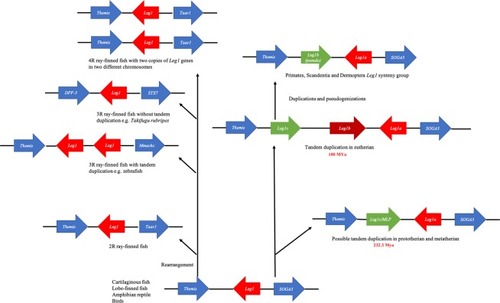
Analysis of the |
|
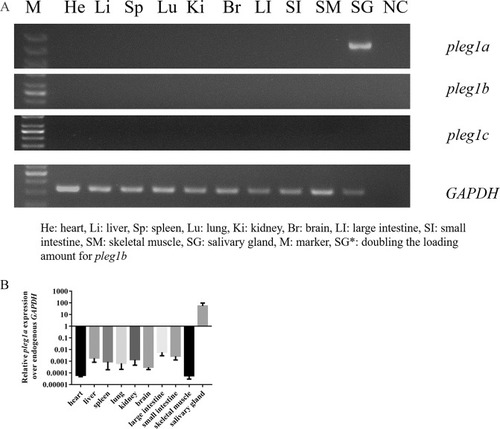
Expression patterns of |
|
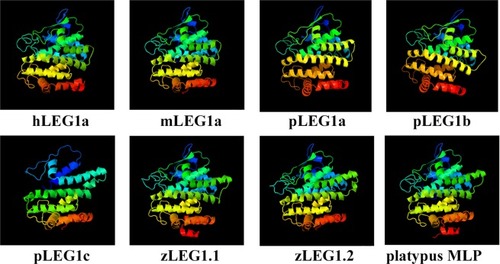
Structural comparison of LEG1 proteins from different species. The platypus MLP protein structure was retrieved from the PDB (4V00), while the others were predicted using Phyre 2. The colors are in rainbow order with red and blue colors indicate the N- and C- termini of LEG1, respectively. All LEG1 proteins exhibit the similar structural prediction result expect for pLEG1c, which is slightly different from others due to the lack of the signal peptide. |
|
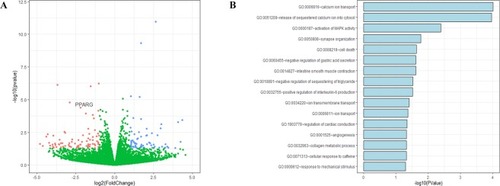
RNA-seq analysis of cells with overexpressing |