- Title
-
Zebrafish C-reactive protein isoforms inhibit SVCV replication by blocking autophagy through interactions with cell membrane cholesterol
- Authors
- Bello-Perez, M., Pereiro, P., Coll, J., Novoa, B., Perez, L., Falco, A.
- Source
- Full text @ Sci. Rep.
|
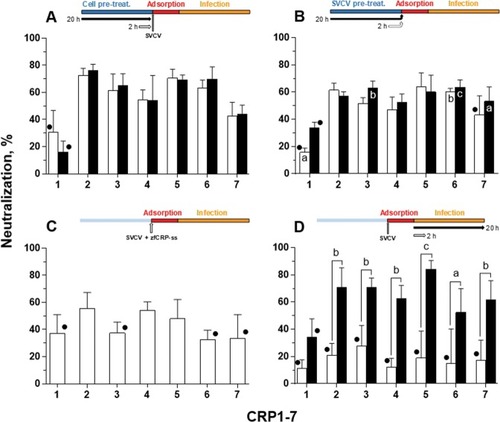
Effect of CRP treatment on SVCV replication in EPC cells. The neutralization activity of CRP1-7 was analysed by adding each CRP at different points of SVCV replication by incubating the CRP with ( |
|
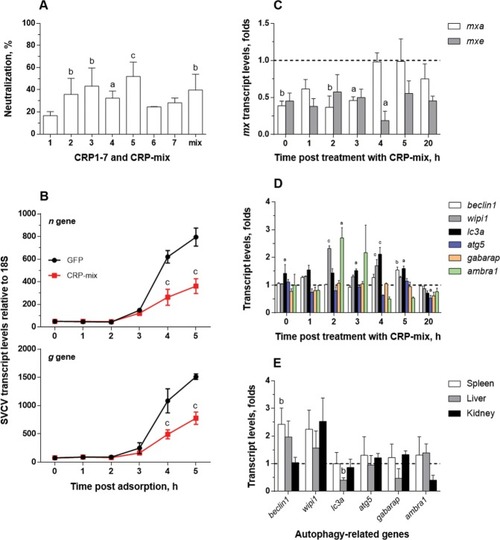
Interaction of CRP1-7 on SVCV replication in EPC cells. ( |
|
Interaction of CRP1-7 on SVCV replication in ZF4 zebrafish cells. ( |
|
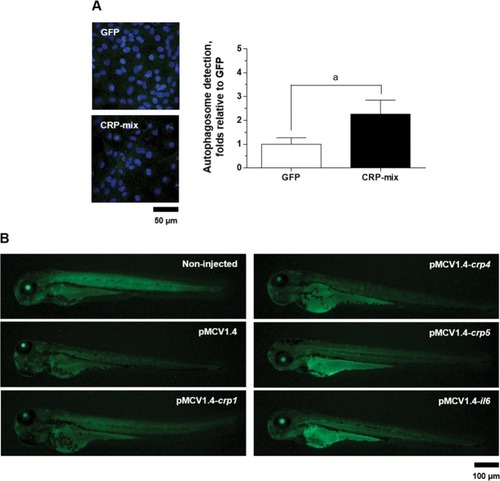
LC3 recruitment by selected CRPs in ZF4 cells and in zebrafish larvae. ( |
|
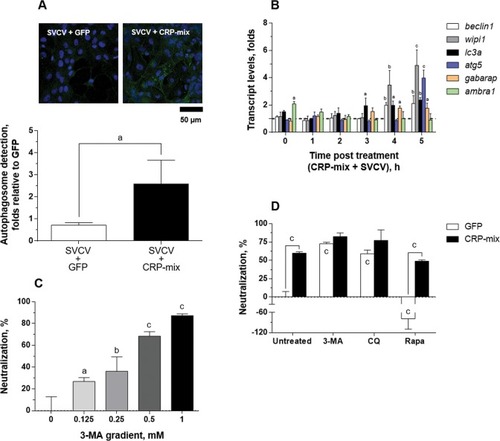
Autophagy induced by CRP-mix on SVCV replication in the ZF4 cells. ( |
|
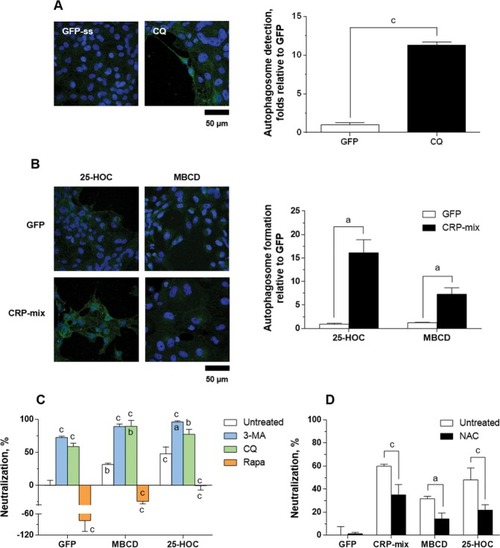
Autophagy and ROS generation during SVCV neutralizing activity induced by 25-HOC and MBCD together with the CRP-mix. Representative confocal images of the FITC immune-labelled LC3B in the ZF4 cells treated with ( |
|
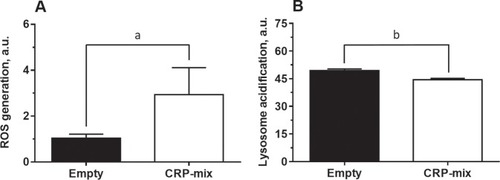
ROS generation and alkalization of intracellular pH induced by CRPs. ( |
|
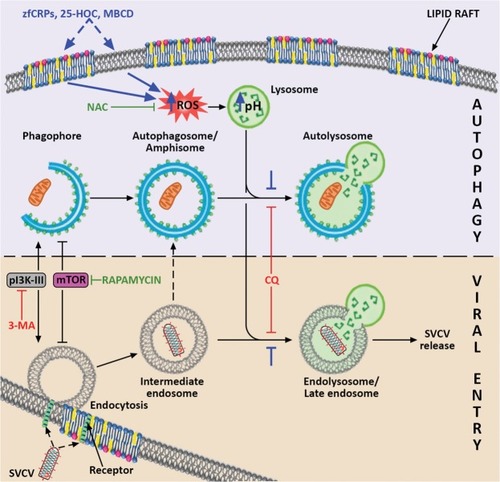
Proposed model for the mechanism by which CRPs, 25-HOC and MBCD interact with autophagy and SVCV entry. It is suggested that these three compounds (their proposed effects are indicated in blue) produce an imbalance in the membrane cholesterol of the lipid rafts, which induces the increase of intracellular ROS. In turn, ROS stimulate the increase in lysosomal pH, which reduces both the fusion of lysosomes and intermediate endosomes (indicated with blue stoppers), and consequently the formation of late endosomes/endolysosomes. Because of their low pH, SVCV requires the formation of endolysosomes to trigger the fusion conformation of the SVCV G protein for viral entry, and a blockade of endolysosomes thus impairs SVCV release into the host’s cytosol. The scheme shows that SVCV endocytic and autophagy pathways share common elements that enable the action of particular autophagy modulators on both of them. The convergence of pathways that may result in the formation of amphisome, as described for other viruses, is also indicated. The positive regulators of both routes are drawn in green, and the negative regulators are presented in red. Artwork drawn and provided by Mr. Diego Sanz. |