- Title
-
Hypoxia Induces Macrophage tnfa Expression via Cyclooxygenase and Prostaglandin E2 in vivo
- Authors
- Lewis, A., Elks, P.M.
- Source
- Full text @ Front Immunol
|
Macrophage |
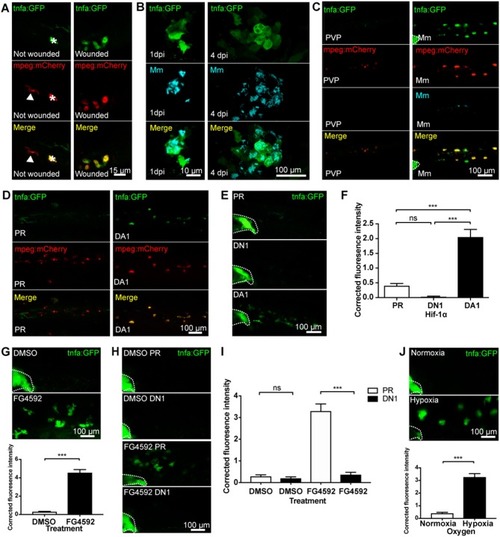
|
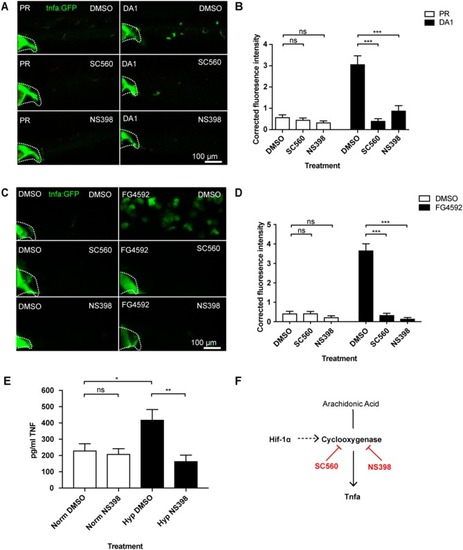
Hif-1α-activated |
|
Injury and infection induced |
|
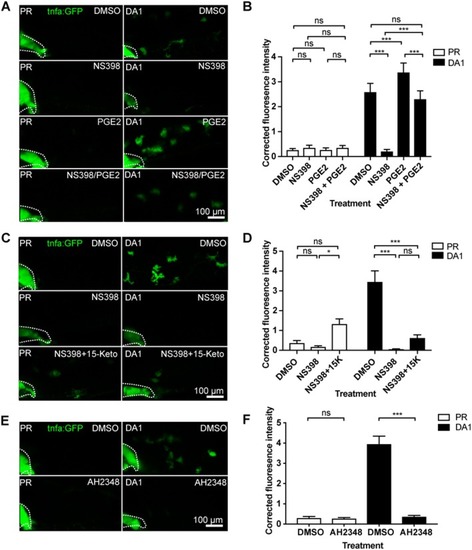
Blocking 15-lipoxygenase or leukotriene B4 receptors does not abrogate DA-Hif-1α-upregulation of |
|
Hif-1α-induced |