- Title
-
Zebrafish as a predictive screening model to assess macrophage clearance of liposomes in vivo
- Authors
- Sieber, S., Grossen, P., Uhl, P., Detampel, P., Mier, W., Witzigmann, D., Huwyler, J.
- Source
- Full text @ Nanomedicine
|
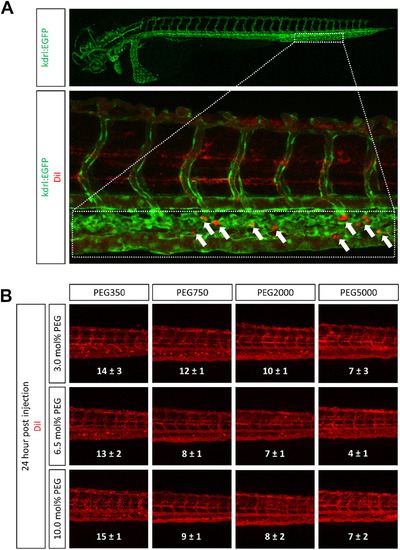
Influence of PEG modification on liposome accumulation in the zebrafish. DiI labeled Liposomes with various PEG molecular weights and PEG densities were injected into blood circulation of zebrafish 2 days post fertilization. (A) The indicated area (dotted box) of injected zebrafish representing part of the caudal vein and the caudal hematopoietic tissue was analyzed for liposome accumulates (white arrows) 24 h post injection. (B) Representative confocal images of the tail region for each liposome formulation are shown. The number of red dots (i.e. liposome accumulates) in the caudal vein was counted and is represented as mean ± SEM (n = 3). |
|
Influence of PEG modifications on macrophage clearance in transgenic zebrafish. DiI labeled liposomes modified with the indicated PEG molecular weights were injected into the blood circulation of zebrafish 2 days post fertilization. Confocal images were acquired 3 and 24 h post injection (hpi). Colocalization analysis of green fluorescent macrophages (KAEDE) and DiI labeled liposomes was performed based on normalized overlap colocalization coefficients. Values are means ± SEM, n = 3. *P < 0.1 or **P < 0.05 (multiple comparison using ANOVA assuming equal variance and Bonferroni correction) as compared to 90 nm sized PEG350 liposomes. |
|
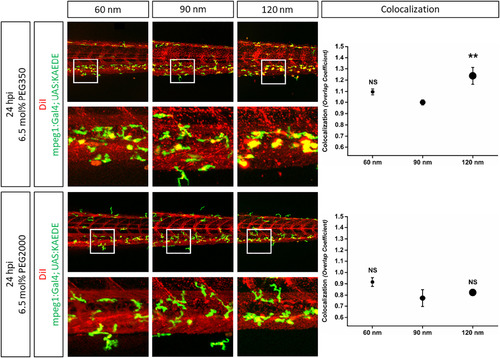
Influence of liposome size and different PEG modification on macrophage clearance in zebrafish. PEG350 or PEG2000 modified and DiIlabeled liposomes with three different sizes (60 nm, 90 nm, and 120 nm) were injected into the blood circulation of zebrafish 2 days post fertilization. Confocal images were acquired 24 h post injection (hpi). Colocalization of green fluorescent macrophages (KAEDE) and DiI labeled liposomes was quantitated using normalized overlap colocalization coefficients. Values are means ± SEM, n = 5. not significant (NS) or **P < 0.05 (ANOVA and Bonferroni correction) as compared to 90 nm sized liposomes. |
Reprinted from Nanomedicine : nanotechnology, biology, and medicine, 17, Sieber, S., Grossen, P., Uhl, P., Detampel, P., Mier, W., Witzigmann, D., Huwyler, J., Zebrafish as a predictive screening model to assess macrophage clearance of liposomes in vivo, 82-93, Copyright (2019) with permission from Elsevier. Full text @ Nanomedicine