- Title
-
Loss of embryonic neural crest derived cardiomyocytes causes adult onset hypertrophic cardiomyopathy in zebrafish
- Authors
- Abdul-Wajid, S., Demarest, B.L., Yost, H.J.
- Source
- Full text @ Nat. Commun.
|
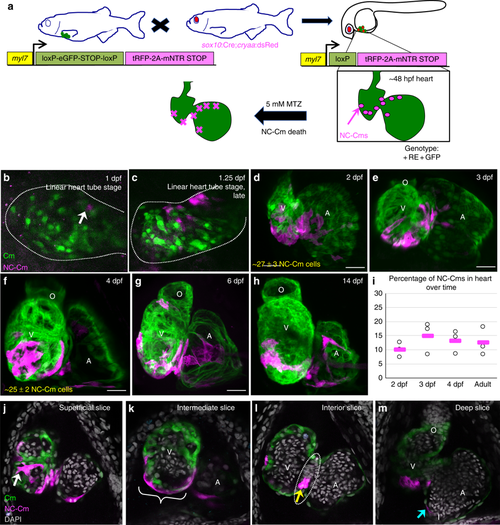
Mapping neural crest-derived cardiomyocytes (NC-Cms) during zebrafish heart development. a Schematic of the NC-Cm lineage labeling and ablation setup using the Cm:KillSwitch transgenic (myl7-driven transgene) crossed to the NC driver Tg(sox10:cre;cryaa:dsRed). Metronidazole (MTZ) treatment causes mNTR-expressing cells to die, i.e., NC-Cms switched to express tagRFP + and mNTR. b–h Contribution of NC-Cms to the developing heart over time. Confocal maximum intensity images of each development stage (1–14 dpf). On average, 27 ± 3 NC-Cms were found at 2 dpf, and this number did not significantly increase by 4 dpf (25 ± 2 NC-Cms). Quantification was from confocal 3D stack images at indicated timepoints and from three individuals. Dotted line outlines heart tube. Scale bar = 25 µm. i Graph displays percent of total cardiomyocytes that are tRFP + (i.e., NC-Cms). Cells were dissociated from isolated hearts at each of the indicated developmental stages and analyzed by flow cytometry. After live cell gating, the sum of GFP and RFP-positive counts was deemed as the total cardiomyocyte count. RFP counts divided by total cardiomyocyte count was used to compute percentages. Circles are biological replicates at each time point and bars are the average of replicates. j–m Confocal slices of a 4 dpf heart from NC-Cm lineage-labeled embryos. White arrow indicates trabeculating NC-Cm. Bracket denotes the apex of the ventricle. Dashed line encircles the AV canal and the yellow arrow indicates the couple of NC-Cms found on the outer curvature of the AV canal. Blue arrow indicates the NC-Cm found at the border of the inflow tract. O = outflow tract, V = ventricle, A = atrium. Scale bar is 30 µm. Images are representative of n ≥ 3 EXPRESSION / LABELING:
Construct:
Tg(myl7:LOXP-EGFP-LOXP-TagRFP-2A-NTR)
|
|
NC-Cm ablation alters trabeculae patterning. a Schematic of NC-Cm ablation protocol. Tg(Cm:KillSwitch) and Tg(sox10:cre;cryaa:dsRed) heterozygotes were crossed to generate three genotypes: Tg(Cm:KillSwitch) ( + GFP); Tg(sox10:cre:cryaa:dsRed) ( + RE); or double-transgenic ( + RE + GFP). Double-transgenic embryos were treated with DMSO (control) or MTZ from 30–48hpf to ablate NC-Cms. Sibling Tg(Cm:KillSwitch (-RE + GFP) were treated with MTZ as a drug control. Embryos were phenotyped at 5 dpf. b–d Confocal maximum intensity projection images from three hearts at 5 dpf from each condition. NC-Cm cells (tagRFP + ) were absent from the MTZ)-treated + RE + GFP embryos compared with their DMSO treated sibling controls (d compared with b). White arrows indicate a remnant, extruding NC-Cm as a consequence of cell death. Scale bar = 100 µm. e Quantification of the number of tagRFP + cells in the 5 dpf ventricle (“internal”) in control (DMSO + RE + GFP) and NC-CM ablated embryos (MTZ + RE + GFP). Bars are mean of individual hearts (open circles) quantified in each condition. Individual protrusions from the ventricle (“blebbs”) that were tagRFP + were also quantified. f Trabeculation analysis of control and NC-Cm-ablated hearts at 5 dpf. Control hearts (left panels) had an array of trabeculae with primary branches arranged along anterior–posterior coordinate. In contrast, NC-Cm-ablated ventricles (from protocol in a) had poorly organized trabeculae (right panels). The angle of the primary branch of a trabecula and relative anterior–posterior position of the primary branch within the ventricle were measured as shown in bottom panel, magenta arrows depict primary trabecula branch; relative position in ventricle axis as represented by bracket. The position and angle of the primary trabeculae branches were measured relative to the AV canal. These data were collected for controls (-RE + GFP, MTZ treated) and NC-Cm-ablated hearts ( + RE + GFP, MTZ treated) and a slope was computed using the trabecula angle to position data for each individual heart (see Supplementary Figure 7). g Computed slope values for individual hearts in each treatment. Mean is indicated by the dashed line and median by the solid line. The slope measurement was significantly different for NC-Cm-ablated hearts compared with their sibling controls. P-values computed by TukeyHSD on ANOVA (F(62,60) = 3.31) EXPRESSION / LABELING:
PHENOTYPE:
|
|
NC-Cm cells regulate Notch signaling during trabeculation. a–f Ventricle sections of a 3 dpf embryo from a sox10:tagRFP transgenic line crossed with the Notch reporter Tp1:GFP line, immunostained for the cardiomyocyte marker MF20. NC-derived lineages (tagRFP) did not show high levels of Notch response (GFP). Arrows indicate trabeculating NC-Cms that were next to a Notch-activated cardiomyocyte (arrowheads). Scale bar = 10 µm. g qPCR gene expression of isolated 4 dpf hearts from NC-Cm-ablated and control siblings. Values are delta delta Ct computations. Bars represent mean of four biological replicates. Points are individual experiments. Delta Ct values used to compute standard T test significance (P-values shown) between NC-Cm ablated and control delta Ct values. h–k Jag2b fluorescent in situ hybridization in a magnified ventricle section of 3 dpf NC-Cm lineage-labeled embryos. Scale bar = 5 µm. Image is representative of N = 17 embryos imaged and analyzed in probe positive in situs compared with probe negative embryos imaged and analyzed. l Quantification of jag2b fluorescent in situ signal in individual NC-Cm and Cm ventricle cells. Relative intensity was computed by the average intensity of all jag2b expression in the heart cells and then comparing with individual NC-Cm tRFP + and Cm GFP + cell intensities. Circles are individual cells from N = 3 embryos from three independent in situ experiments. Bar = mean. Standard T test was used for P-value. m Schematic model in which NC-Cms provide spatial patterning of trabeculation using Notch signaling components. During the transition to initiate trabeculae, NC-Cms provide a significant source of jag2b expression that triggers its neighbor Cm to repress protrusion and trabeculation initiation. This results in evenly, spatially distinct trabeculae branches at 5dpf. However, without NC-Cms and their major source of jag2b expression (although not exclusive as indicated by the remaining Cm expressing jag2b at 3dpf), more Cms are poised to protrude and create trabeculae, yielding conjoined branches and poor trabeculae spacing by 5 dpf EXPRESSION / LABELING:
PHENOTYPE:
|
|
Ablation of embryonic NC-Cms results in adult-onset hypertrophic cardiomyopathy and heart failure. a–c Whole-mount fluorescent images of adult hearts from embryonic NC-Cms ablation experiments (protocol in Fig. 2a). Red numbers represent percent of NC-Cms relative to total cardiomyocytes (GFP + and RFP + ), not total heart cells, quantified by flow cytometry (FACS) of dissociated adult hearts (Supplementary Figure 9). d FACS quantification of NC-Cms. Dots are individual hearts from each condition and bars = mean of individuals in each group (n = 4 + RE + GFP, DMSO and n = 5 + RE + GFP, MTZ) e–g Fluorescent microscopy sections of hearts from sibling individuals as in A-C. scale bar = 100 µm. h Quantification of area of ventricle covered in cardiomyocytes from sections similar to d–f. Dots represent sibling individuals from each condition pooled from biological replicates. Solid line = median, dashed line = mean. P-values computed by TukeyHSD on ANOVA (F(2,16) = 9.48). i-j Examples of single-cell cardiomyocyte morphology from dissociated control (i) and NC-Cm-ablated adult ventricles. Cells were cultured in chambers for 24 h to allow cell attachment, then fixed and stained with anti-GFP and DAPI and visualized by microscopy. Scale bar = 10 µm. k Quantification of individual cardiomyocyte area from chamber cultures as in “i, j”; n ≥ 25 cells per sample. P-value from standard T test. l Quantification of individual cardiomyocyte lengths from chamber cultures as in “i, j”. P-value from standard T test. m Example still image from movies of swim tunnel assays of individual male zebrafish. Red arrow indicates water current direction. n Swim trial assay provided incremental water speed increases of 0.05 m/s every 6 min (solid black line). Average assay results for DMSO control (light green, n = 4), MTZ control (dark green, n = 5) and NC-Cm-ablated (magenta, n = 8) adults. Data are amalgamated from n = 5 biological replicates. See Supplementary Movie 2. o Individual Ucrit results normalized to body length from assay in "m". See Methods for Ucrit calculation. Dots represent individual males from each condition. Solid line = median, dashed line = mean. P-values computed by TukeyHSD on ANOVA (F(2,13) = 24.65) PHENOTYPE:
|
|
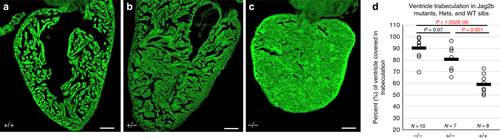
Jag2b mutant adults phenocopy hypertrophic cardiomyopathy of the NC-Cm ablated adults. a–c Microscopy sections of adult ventricles from jag2b wild-type ( + / + ), heterozygous ( + /-), and homozygous (-/-) mutants. Sections were stained with phalloidin-488 and imaged. Scale bar = 100 µm. d Area of ventricle with 488 + trabecular myocardium was quantified as in 4 H and tested for significance by two-way T tests PHENOTYPE:
|
|
Cm:KillSwitch transgenic embryos. Whole mount fluorescent images of 4dpf Cm:KillSwitch transgenic embryos showing exclusive GFP fluorescence in the heart. Scale bar = 200px. |
|
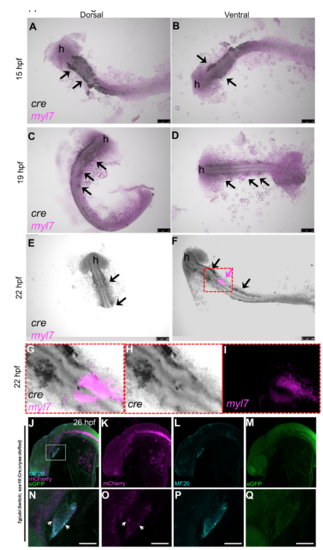
Tg(sox10:Cre;cryaa:dsRed) expression relative to the developing heart. A-I) 15hpf, 19hpf and 22hpf embryos from the Tg(sox10:Cre;cryaa:dsRed) line were probed for myl7 and cre RNA detection by two color in-situ hybridization. myl7 was detected by FastRed staining (fluorescence in Red channel, magenta) and cre by NBT-BCIP stain (black). Arrows point to cre expression in the stereotypical neural crest lineage pattern progressing medially from the dorsal neural tube. Magenta arrow demonstrates the first detection of the myl7+ heart field in the ventral side of 22hpf embryos (low level magenta in A-D was equivalent to background and negative control stained embryos). A few black cre+ cells can be seen nearby the myl7 staining at this stage (black arrows). ‘h’= head region of embryo. Yolks were removed to mediate visualization. Scale bar = 100uM. Right panels are the ventral side images of the same embryos in left panels. G-I) Higher magnification of heart field, from red box shown in F. H and I show separate channels for cre and myl7, respectively. J-Q) 26hpf embryos from a cross of Tg(sox10:Cre;cryaa:dsRed) to the Tg(ubi:Switch) line were stained for MF20 and imaged for GFP, mCherry and MF20. The bottom panels are close up images of the heart area indicated by white dashed box in J. mCherry signal indicates the Cre switched neural crest cells. A few mCherry positive cells overlap with the MF20 heart marker indicating the first appearances of NC-Cms (white arrows). Scale bar = 75uM in prime labelled panels. |
|
A) Cardiomyocyte labelling of Tg(Cm:KillSwitch) crossed to Tg(sox10:Cre;cryaa:dsRed). 5dpf embryos were immunostained with anti-sarcomere myosin heavy chain antibody MF20 and imaged in conjunction with tagRFP and GFP fluorescence. Confocal slices of deep (top panel) and superficial (bottom panel) areas of the heart are shown. Arrows point to examples of MF20 stain overlap with the NC-Cm tagRFP fluorescence. Scale bar = 50uM. B) Quantification of the percent of NC-Cms found in trabeculae of the ventricle relative to total ventricle NC-Cms. tagRFP+ NC-Cms were counted from 4 and 5dpf ventricles and designated as being within a trabeculae or not. The number found within trabeculae was divided by the total NC-Cms in the ventricle to yield percentage contribution. Circles represent individual hearts that were counted and bars represent mean of individual data points. |
|
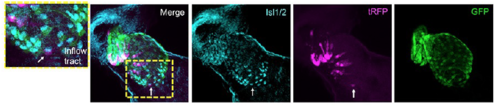
Isl1/2 antibody of anterior second heart field and lineage-labeling of NC-Cms indicates interdigitation of neural crest cells into anterior second heart field. Arrow indicates single NCCm in the inflow tract that is positive for Isl1/2, also a marker of pacemaker cells1. tagRFP and GFP were detected by immunolabeling. |
|
Cell death in NC-Cms after MTZ treatment. Embryos derived from transgenic crosses as described in Figure 1 and 2A. A) Control: DMSO treated double transgenic siblings. B) Control: MTZ treated single transgenic siblings. C) MTZ treated double transgenic (+RE+GFP) had positive staining for Active Caspase 3, indicative of cell death (arrows), that was not observed in hearts from controls. D) Quantitation of active Caspase staining signal in control and ablated embryos at 55hpf. Bars indicate mean of n=3 individual heart images (open circles). Red dashed line indicates background level of fluorescent signal in detection channel for active Caspase (179 RFU). |
|
Jag2b fluorescent in-situ hybridization in NC-Cm labelled embryos. 3dpf hearts from NC-Cm labelled embryos were subject to the fluorescent in-situ protocol with and without jag2b-TRITC probe as well as immunostaining for tRFP and GFP. Remnant RFP expression from the NC-Cms is present in the tritc channel however distinct from jag2b probe detection (white arrows and astericks compared to no-probe controls). White arrows show NC-Cm specific colocalization of jag2b signal and asterisks show other Cm expression or non-Cm expression. The strong tail expression of jag2b was used as a positive control for the fluorescent in-situ hybridization experiment. |
|
NC-Cm labeling in adult transgenic fish. A) Flow cytometry analysis and quantification of labeled cardiomyocytes in Cm:KillSwitch x Tg(sox10:cre;cryaa:dsRed) individual 15 adult heart versus AB control. GFP and RFP gates were drawn based on non-transgenic/non-fluorescent AB wild-type hearts. Dissociated cells were gated on viability (DAPI) and singlets before analyzing GFP and RFP populations. Three or more individual hearts were similarly analyzed by flow cytometry to quantify numbers of GFP+ and RFP+ cardiomyocytes. GFP+ and RFP+ percentages were added to quantify total number of cardiomyocytes in a whole heart and then RFP+ numbers were divided by this to generate their percent contribution to the total adult cardiomyocyte population. The average values of these quantifications are listed in Figure 4A and C. Heart dissociation protocol was carried out as previously described2. B-D) Adult sections of ventricles from control (+RE+GFP, DMSO). Sections were stained with anti-tagRFP and anti-GFP antibody. Fluorescent images were captured at 20X magnification. Yellow arrows point to tagRFP+ NC-Cms in trabeculae of adult heart. E-G) Higher magnification of tagRFP+ trabeculae from control hearts as in B-D. Scale bar =50uM H-J) NC-Cm ablated (+RE+GFP, MTZ) heart sections and example of remnant tagRFP (yellow arrow) seen in the NC-Cm ablated ventricle sections. Scale Bar = 200uM. |
|
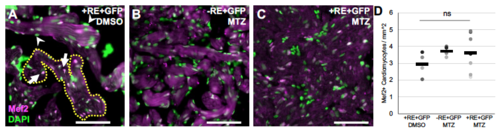
Quantification of cardiomyocyte number and cell size in adult hearts. A-C) Examples of microscopy sections of ventricles stained with Mef2 and DAPI. Scale bar =15uM. Nuclei that are both Mef2 and DAPI positive (arrows in A) were used to count the number of cardiomyocytes per area defined by a ROI selection of the auto fluorescent trabecular myocardium, an example ROI selection is shown by yellow dashed line. Non-cardiomyocyte nuclei that were green only (arrowheads in A) were not counted. D) Cardiomyocyte numbers quantified from microscopy sections. Dots represent individual adult section measurements and bars the mean of each sample. ns = not significant from standard two-way T-test. |