- Title
-
Myotonia Congenita-Associated Mutations in Chloride Channel-1 Affect Zebrafish Body Wave Swimming Kinematics
- Authors
- Cheng, W., Tian, J., Burgunder, J.M., Hunziker, W., Eng, H.L.
- Source
- Full text @ PLoS One
|
Characterization of stable transgenic zebrafish embryos using Tg (α-actin:hCLCN1I553F/H555N-EGFP). (A) A pool of 28 h dpf embryos from the stable transgenic founder fish showing the EGFP expression. Nonfluorescent embryos are nontransgenic siblings. (B) Embryos with positive EGFP expression show the muscle-specific expression in their trunk region. (C) Confocal microscopic image of the trunk of an F1 generation Tg (α-actin:hCLCN1I553F/H555N-EGFP) embryos at 3 dpf. EGFP expression was observed within the zebrafish muscle fibers. (D) Transgenic founders were confirmed by transmission of the CLCN1 transgene to F1 progeny by PCR. Transgenic founders were crossed with WT zebrafish, individual F1 embryos were lysed and PCR were performed using hCLCN1 specific primers and the PCR product analyzed by agarose gel electorphorsis. EGFP-positive embryos are identified based on the presence of the 1.5 kb band (e3, 5, 6, 9 and 10), while the EGFP-negative embryos lack this band (e1, 2, 4, 7, 8, 11 and 12). e; embryo. (E) Transgenic founders were confirmed by Western blot. Protein was extracted from F1 embryos at 2 dpf. Lane 1: WT AB Zebrafish, Lane 2-4: Individual stable transgenic fish expressing the 130 kDa Flag-tagged hCLCN1I553F/H555N-EGFP (see Materials and Methods). Actin was used as a loading control. (F) Transgenic zebrafish embryos at 3 dpf showing low, moderate or high expression of hCLCN1I553F/H555N-EGFP based on GFP intensity. (G) Quantification of hCLCN1I553F/H555N-EGFP expression and classification. For each transgene, 3 siblings of 3 dpf embryos were analyzed. Average pixel intensities (api) for EGFP fluorescence were determined on 5 areas of the trunk and the average for each sibling is plotted. Expression levels of the transgene were arbitrarily classified into low (<10 api), moderate (10-20 api) or high (>20 api). Similar results were obtained for the other transgenes (data not shown). EXPRESSION / LABELING:
|
|
The comparison of averaging spatial-temporal body curvature profiles and averaging temporal tail offset profiles. For each zebrafish, 16-second video sequences at high frame rates were recorded. The video clip was manually split into smaller clips containing one full body waving cycle. The fish body was tracked and the bending degree determined using body curvature and tail offset as criteria. (A-G) averaging spatial-temporal body curvature profiles that are obtained from the following zebrafish: wild-type, hCLCN1-EGFP, hCLCN1L844F-EGFP, hCLCN1I553F/H555N-EGFP, hCLCN1-IRES-EGFP, hCLCN1L844F-IRES-EGFP, hCLCN1I553F/H555N-IRES-EGFP, respectively. We consolidated body curvature measurements calculated at each frame into a matrix. Each column represents the body curvature measurements in one frame. The number of columns is equal to the number of frame of one body waving cycle. We further averaged matrices over all body waving cycles of all zebrafish within same category to obtain an averaging body curvature profile. Red color indicates higher curvature value and larger bending of fish body. The Y-axis represents the body curvature measurements in one frame. The X-axis represents frame index of one video segment of one body waving cycle. (H-I) Consolidated tail offset measurements calculated at each frame into a vector. The vectors are averaged over all body waving cycles of all zebrafish within same category to obtain an averaging tail offset profile. Larger tail offset value indicates smaller bending of fish body. PHENOTYPE:
|
|
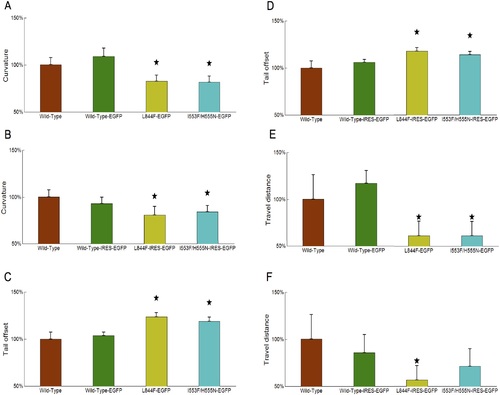
Comparison of body curvature, tail offset and swimming distance measurements between wild-type, control and transgenic zebrafish. We selected the largest body curvature value (A-B); smallest tail offset value (C-D), and travel distance (E-F) from each body waving cycle. All measurements are first averaged over all video clips within same category. The averaged measurements are further divided by the mean measurement of wild-type zebrafish. The measurements are plotted as mean +/ standard deviation. The star symbol in the figure indicates a significant difference (p<0.05) in the Student-t test. PHENOTYPE:
|