- Title
-
Sequencing of pax6 Loci from the elephant shark reveals a family of pax6 genes in vertebrate genomes, forged by ancient duplications and divergences
- Authors
- Ravi, V., Bhatia, S., Gautier, P., Loosli, F., Tay, B.H., Tay, A., Murdoch, E., Coutinho, P., van Heyningen, V., Brenner, S., Venkatesh, B., and Kleinjan, D.A.
- Source
- Full text @ PLoS Genet.
|
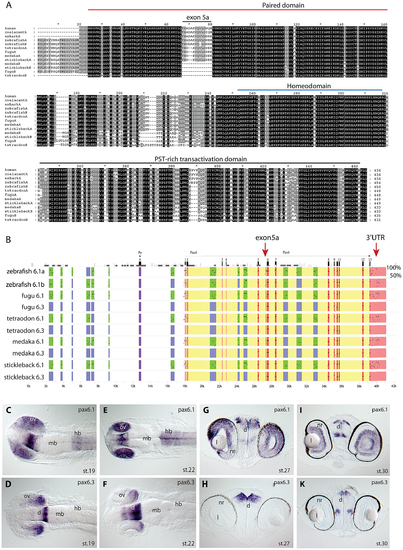
Pax6 gene duplicates in teleost fish. A) Alignment of Pax6 proteins. Amino acid sequences of duplicate Pax6 proteins from various fish species are aligned along those from a variety of tetrapods and elephant shark. Pax6 proteins are highly conserved across vertebrates, particularly in the paired, homeo- and transactivation domains. The potential to encode an N-terminal protein extension is present in all fish Pax6 genes. The alternative exon 5a is present in the Pax6 genes of tetrapods, in one of the Pax6 duplicates in fish, as well as in the second zebrafish Pax6 gene. The other Pax6 genes in the acanthopterygian teleosts lack the alternative exon 5a. B) Percentage Identity Plot (PIP) showing multispecies sequence comparison of the genomic region around the Pax6 transcription unit using human PAX6 locus as baseline sequence. The plot highlights a number of features that indicate different evolutionary origins of the duplicated Pax6 genomic loci in fish species. While strong conservation of exonic sequences (red boxes indicate their positions, black lines/dots show the level of conservation) is seen for the duplicate genes in all fish species, a conspicuous absence of conserved elements is observed in the upstream and intronic regions of the second Pax6 loci of medaka, stickleback, fugu and Tetraodon. In contrast, in zebrafish both duplicate loci contain a largely overlapping array of conserved non-coding elements (CNEs). CNEs are highlighted by green boxes, while their absence is shown by blue boxes. Exon 5a, an alternatively spliced exon located immediately upstream of exon 6 (red arrow), is present in both zebrafish Pax6 loci (Pax6.1a and b), but absent from the second Pax6 loci (Pax6.3) of the other teleosts (note the absence of black dots/lines in the red box for the exon5a position). Similarly, the gene has conserved sequences in its 32UTR (red arrow) that are present in all canonical Pax6 (Pax6.1) loci, and are subpartitioned between the zebrafish Pax6 duplicates, but are not found in the 3′UTRs of the Pax6.3 gene of other fish species. C–K) RNA in situ analysis of medaka Pax6 genes during early embryonic stages: C) At stage 19 expression of medaka Pax6.1 is seen in the optic vesicle (ov), diencephalon (d) and hindbrain (hb), but is absent from the midbrain (mb). D) Expression of Pax6.3 at the same stage is seen in the distal part of the optic vesicle and in the posterior diencephalon. E) At stage 22 Pax6.1 expression is seen in the optic cup (oc), diencephalon and hindbrain, while F) Pax6.3 signal is maintained in the posterior half of the optic cup and has increased in the diencephalon. G) A cross section at stage 27 shows Pax6.1 expression in the neuroretina (nr) and lens (l) epithelium of the eye, and in the dorsal diencephalon. H) In contrast Pax6.3 signal is no longer seen in the retina, but strong expression is maintained in the dorsal diencephalon. I) By stage 30 medaka Pax6.1 expression is restricted to the lens epithelium, the ganglion cell layer and the inner nuclear layer in the retina, and in the dorsal and medial diencephalon and ventral nerve tracts. K) Pax6.3 expression is limited to the dorsal and medial diencephalon. |
|
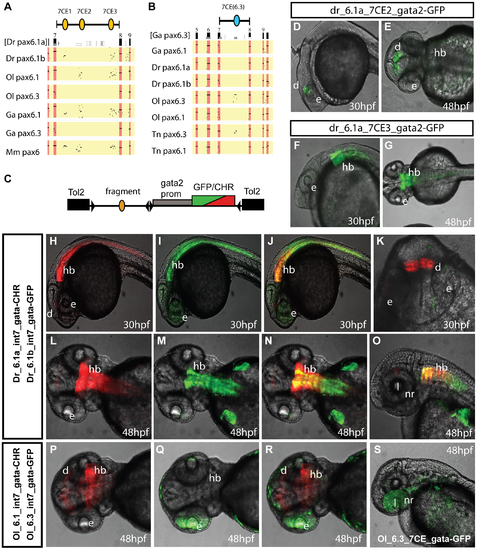
Comparison of functional activity driven by zebrafish and medaka intron 7 sequences. A, B) Comparison of full Pax6 intron 7 sequences of multiple teleost species visualized as PIP plots. A) PIP plot with zebrafish Pax6.1a intron 7 as baseline sequence against intron 7 sequences of the Pax6.1 and Pax6.3 genes of zebrafish (Danio rerio (Dr)), medaka (Oryzias latipes (Ol)), stickleback (Gasteroteus aculeatus (Ga)), tetraodon (Tetraodon nigroviridis (Tn)) and mouse (Mus musculus (Mm)), showing lack of sequence conservation of Pax6.1 intron 7 with the Pax6.3 loci. A variable subset of CNEs has been conserved in the Pax6.1 loci. B) PIP plot using stickleback intron 7 as base sequence against the intron 7 regions of the Pax6.1 and Pax6.3 loci from multiple teleosts, showing the presence of a conserved element unique to the Pax6.3 genes only. C) To assess whether patterns of sequence conservation are reflected at the functional cis-regulatory level the full intron 7 sequences from zebrafish Pax6.1a (Dr6.1a_int7), zebrafish Pax6.1b (Dr6.1b_int7), medaka Pax6.1 (Ol6.1_int7) and medaka Pax6.3 (Ol6.3_int7) were cloned in front of a gata2 minimal promoter-reporter cassette in a Tol2-2way reporter vector system. Most fragments were cloned with both GFP and mCherry as fluorescent reporter to allow combinatorial analysis in dual fluorescence reporter transgenic zebrafish. (D–G) In addition smaller fragments containing the individual 7CE1, 7CE2 and 7CE3 elements from the zebrafish Pax6.1a gene, and the 7CE(6.3) element from the medaka Pax6.3 gene were also cloned and used to produce transient transgenic zebrafish. (D, E) Lateral and dorsal views of transgenic fish for the Dr6.1a_7CE2 element show expression in the diencephalon (d) at 30 and 48 hours post fertilization (hpf). (F, G) Lateral and dorsal views of transgenic fish for the 7CE3 element of zebrafish Pax6.1a show expression in the hindbrain (hb) at 30 and 48 hpf. (H–O) Expression driven by the full intron 7 sequences of zebrafish Pax6.1a and Pax6.1b is consistent with the presence or absence of the 7CE2 and 7CE3 elements. mCherry fluorescence at 30 and 48 hpf recapitulates the combined pattern of the 7CE2 and 7CE3 elements with expression seen in hindbrain (hb) and diencephalon (d). GFP fluorescence, driven by zebrafish Pax6.1b intron 7 is observed in the hindbrain and neural tube of transgenic fish, but no signal is seen in the diencephalon in accordance with the absence of the 7CE2 element from the Dr6.1b intron7. A difference in the detail of hindbrain/neural tube expression driven by the Dr6.1a and 6.1b intron 7 sequences is also observed with a stronger and wider expression of Dr6.1a_int7 in the hindbrain and decreasing towards the caudal neural tube, while Dr6.1b_int7 driven expression is narrower in the hindbrain but maintained more evenly along the neural tube. l, lens; nr, neuroretina. (P–S) Medaka Pax6.1 intron 7 drives expression in a similar pattern to zebrafish Pax6.1a with clear expression in diencephalon and hindbrain, with decreasing levels in the neural tube, in accordance with the conservation of the 7CE2 and 7CE3 elements in the intron. Medaka Pax6.3 intron 7 (Ol6.3_int7) drives GFP reporter expression in the eye (e) of transgenic zebrafish, and this expression is replicated when the 7CE(6.3) element, conserved only in Pax6.3 loci, is used on its own. |
|
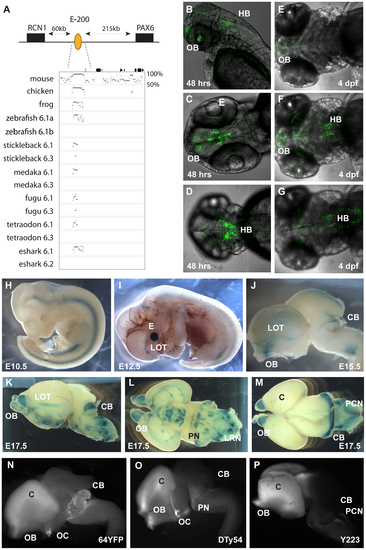
Characterization of the E-200 CNE in mouse and zebrafish reporter transgenics. A) The E-200 CNE is a deeply conserved cis-element found upstream of all vertebrate Pax6.1 loci and located about 200 kb upstream of human Pax6. A shorter stretch of the element is also conserved in the Pax6.3 loci of fugu and stickleback. (B–G) Fluorescent reporter transgenic zebrafish with the human E-200 element. B) Lateral view of an E-200-gata2-GFP reporter line showing expression in the olfactory bulbs (OB) and hindbrain (HB). C) Confocal image of the ventral head region of a 48 hpf transgenic fish reveals expression in the olfactory bulbs and the proliferative zones of the ventral brain region. D) Confocal view of the dorsal embryo head showing expression in the hindbrain. (E–G) Ventral, medial and dorsal confocal views show GFP signal in the olfactory bulbs, lateral olfactory tracts, forebrain (E, F) and hindbrain (F, G) of a 4 dpf reporter transgenic fish. (H–P) LacZ reporter transgenic mice with the human E-200 element. (H) At E10.5 no enhancer specific expression is observed. Signal in the neural tube is intrinsic to the hsp68-LacZ cassette, and in the genital ridge is aspecific. I) From E12.5 expression starts to appear in the olfactory tracts at the base of the cortex. J) At E15.5 staining is seen in the olfactory bulbs (OB), the lateral olfactory tracts (LOT) and a thin band in the dorsal cerebellum (CB). Expression is also seen in the precerebellar neuro-epithelium (PCN), migratory streams and the lateral reticular nuclei (LRN) of the hindbrain. K) This pattern is maintained at E17.5, with increased cerebellar expression covering the full width of the organ. L) Ventral view of an E17.5 dissected brain shows expression in the lateral olfactory tracts and in the precerebellar nuclei including the pontine gray nuclei (PN). Staining in hypothalamic areas was not seen in other transgenic lines, while expression in the olfactory bulbs varied in strength between lines. M) Dorsal view of an E17.5 brain shows the expression in the cerebellum and precerebellar neuroepithelium. N–P) Comparison of reporter YAC transgenic mice carrying 420 kb of the human PAX6 locus, which does not extend to include the E-200 long-range enhancer, to a targeted reporter insertion into the endogenous Pax6 locus shows a deficiency of reporter expression in the cerebellum of the YAC reporter lines. N) Fluorescence of a YFP reporter integrated into the endogenous mouse Pax6 gene is found in the olfactory bulbs (OB), cortex (C), optic chiasm (OC), cerebellum (CB) and pontine migratory stream (PMS). While reporter fluorescence signal of comparable strength is seen in both (O) single copy or multi-copy (N) YAC transgenic lines carrying 420 kb of the human PAX6 locus in the olfactory bulbs, cortical lobes, optic chiasma and pontine migratory streams, expression is absent from the cerebella of these transgenics. |
|
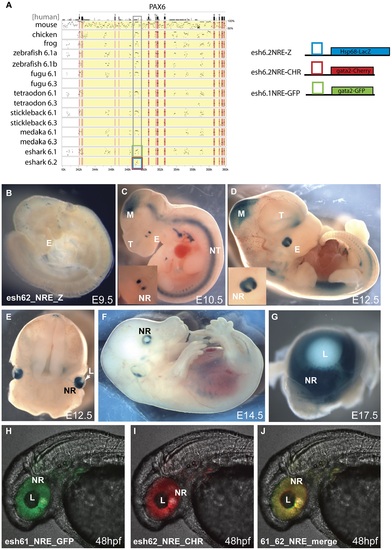
Transgenic analysis of the elephant shark Pα promoter/neuroretinal enhancer (NRE) element. A) PIP plot of the Pax6 intragenic region shows the conservation at the P±/NRE in intron 4 of mammalian Pax6 in both Pax6 loci in elephant shark. The NRE(Cm6.2) element is the only conserved non-exonic sequence between the Pax6.1 and Pax6.2 loci. To allow comparison with the mouse neuroretinal enhancer (NRE) LacZ reporter transgenic mice were made with the Cm6.2 element. (B-G) LacZ staining is seen in the neuroretina of these mice from E10.5 in a pattern that is highly similar to expression driven by the mouse element. B) No expression is seen at E9.5. C) Expression starts at E10.5 at the lateral sides of the developing eyes (E). Expression in the midbrain (M) is due to site of integration and is not seen in other lines. D) By E12.5 expression has widened to include the whole retina. The inset shows a larger view of the eye. E) Cross section through a E12.5 embryo head shows staining in the neuroretina (NR) and optic nerve. F) Expression continues at E14.5 in the neuroretina, and G) is maintained at E17.5 in the retina but not in the lens. (H-J) To test for functional conservation of the elephant shark Pax6.1 and Pax6.2 NRE we assessed the elements in zebrafish transgenics using the dual colour reporter system. H) The NRE element from the elephant shark Pax6.1 locus drives GFP reporter expression in the retina of transgenic zebrafish at 48 hpf. I) The NRE element from the elephant shark Pax6.2 locus similarly drives mCherry in the neuroretina at 48 hpf. J) The merged image shows functional equivalence of the Cm6.1 and Cm6.2 NRE elements in driving reporter expression in the zebrafish retina. NR, neuroretina, L, lens. EXPRESSION / LABELING:
|
|
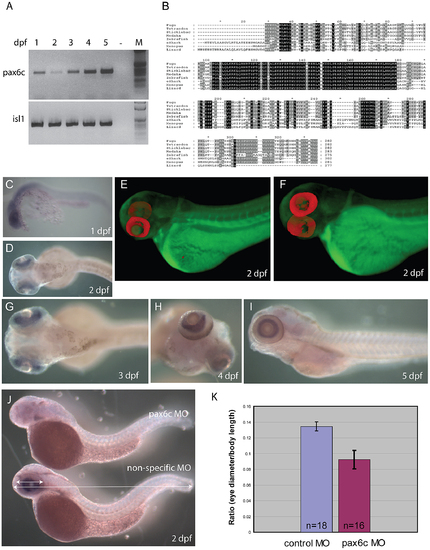
Characterization of zebrafish Pax6.2a. A) rtPCR for Pax6.2a using cDNA from one to five dpf zebrafish embryos showing expression of the gene at all stages. rtPCR using Islet-1cDNA was used as control. B) Protein alignment of Pax6.2 from multiple species showing homology between the Pax6.2 proteins. Homology is highest in the homeobox but extends to several motifs in the PST domain. Zebrafish Pax6.2a shares highest homology with Pax6.2b proteins from other teleosts. (C–I) RNA in situ hybridisation analysis on fixed zebrafish embryos from 1 dpf to 5 dpf. C) At 24 hpf staining is seen in the head region of embryos. D) By 48 hpf expression has become limited to the developing retinae. E,F) Optical Projection Tomography (OPT) stills reveal the restricted expression of Pax6.2 in the retina where it appears limited to the inner nuclear layer. G–I) Expression in the inner nuclear layer of the retina is maintained at 3 dpf (G), 4 dpf (H) and 5 dpf (I). J) Morpholino depletion of Pax6.2a gives rise to zebrafish embryos with relatively smaller eyes compared to embryos injected with a control morpholino. Embryos were fixed at 2 dpf and tested for the presence of Pax6.2 transcript by in situ hybridization. K) Quantification of eye size reveals a 30% reduction in eye size relative to total body length in Pax6.2a morphants versus control morpholino injected embryos. |
|
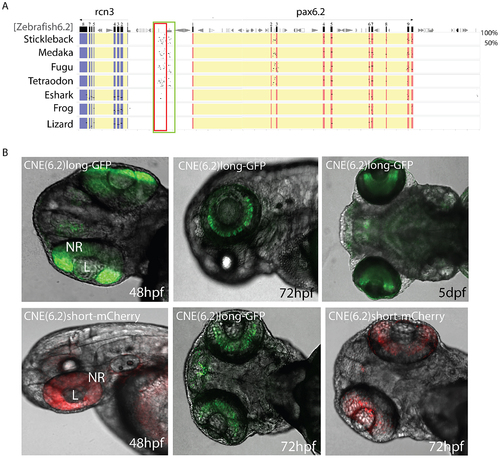
Identification of a novel enhancer specific for the inner nuclear layer of zebrafish retina. A) Sequence alignment of the Pax6.2 genomic region with the Pax6.2 loci from multiple species reveals a region of clear sequence conservation upstream of the gene. A wide fragment of homology is found between zebrafish Pax6.2 and the acanthopterygian fish, with a smaller stretch of conservation to the elephant shark Pax6.2 locus. No conservation is seen with the frog and lizard loci. B) The zebrafish Pax6.2 CNE was cloned as a longer fragment covering the teleost conservation and as a shorter fragment centered on the elephant shark conserved sequence. Dual colour fluorescence transgenesis with both fragments produced a highly specific expression pattern in the retina at 48 hpf, and became restricted to the inner nuclear layer of the retina at 72 hpf and 5 dpf. NR, neuroretina, L, lens. |
|
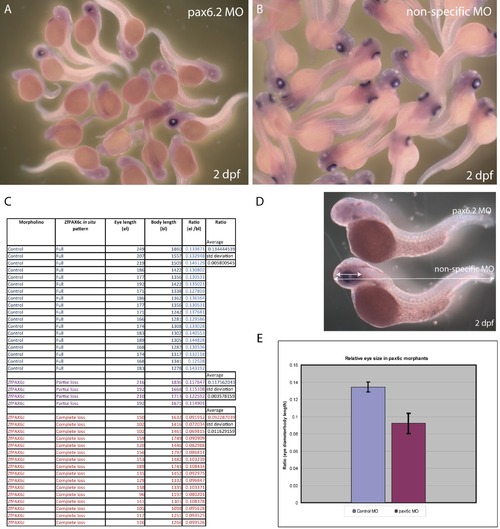
Depletion of Pax6.2 by morpholino knock-down. A) a Pax6.2 morpholino. and B) a control morpholino were Injected into zebrafish oocytes. Pools of embryos were fixed at 2 dpf and tested for the presence of Pax6.2 transcript by in situ hybridization. Pax6.2 ISH signal was absent or greatly reduced in the majority of pax6.2 morpholino injected embryos, but was unaffected by control morpholino injections. C, D) Diameter of the eye was measured relative to the total body size of the embryos, and the average ratio of eye diameter versus body length was calculated. E) Chart showing the eye diameter of pax6.2 morphants was on average 30% reduced compared to embryos injected with a non-specific morpholino. |