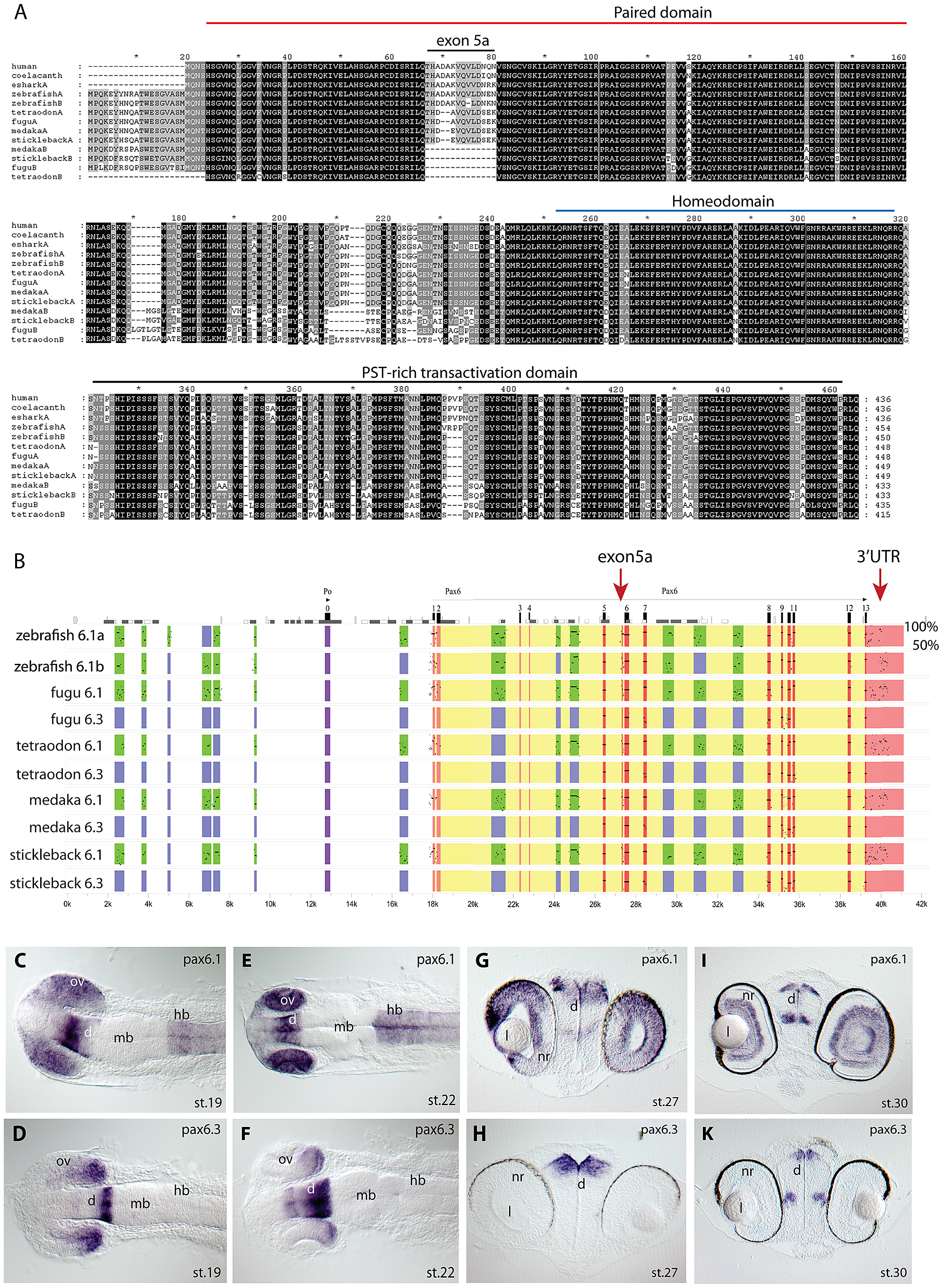
Fig. 1 Pax6 gene duplicates in teleost fish.
A) Alignment of Pax6 proteins. Amino acid sequences of duplicate Pax6 proteins from various fish species are aligned along those from a variety of tetrapods and elephant shark. Pax6 proteins are highly conserved across vertebrates, particularly in the paired, homeo- and transactivation domains. The potential to encode an N-terminal protein extension is present in all fish Pax6 genes. The alternative exon 5a is present in the Pax6 genes of tetrapods, in one of the Pax6 duplicates in fish, as well as in the second zebrafish Pax6 gene. The other Pax6 genes in the acanthopterygian teleosts lack the alternative exon 5a. B) Percentage Identity Plot (PIP) showing multispecies sequence comparison of the genomic region around the Pax6 transcription unit using human PAX6 locus as baseline sequence. The plot highlights a number of features that indicate different evolutionary origins of the duplicated Pax6 genomic loci in fish species. While strong conservation of exonic sequences (red boxes indicate their positions, black lines/dots show the level of conservation) is seen for the duplicate genes in all fish species, a conspicuous absence of conserved elements is observed in the upstream and intronic regions of the second Pax6 loci of medaka, stickleback, fugu and Tetraodon. In contrast, in zebrafish both duplicate loci contain a largely overlapping array of conserved non-coding elements (CNEs). CNEs are highlighted by green boxes, while their absence is shown by blue boxes. Exon 5a, an alternatively spliced exon located immediately upstream of exon 6 (red arrow), is present in both zebrafish Pax6 loci (Pax6.1a and b), but absent from the second Pax6 loci (Pax6.3) of the other teleosts (note the absence of black dots/lines in the red box for the exon5a position). Similarly, the gene has conserved sequences in its 32UTR (red arrow) that are present in all canonical Pax6 (Pax6.1) loci, and are subpartitioned between the zebrafish Pax6 duplicates, but are not found in the 3′UTRs of the Pax6.3 gene of other fish species. C–K) RNA in situ analysis of medaka Pax6 genes during early embryonic stages: C) At stage 19 expression of medaka Pax6.1 is seen in the optic vesicle (ov), diencephalon (d) and hindbrain (hb), but is absent from the midbrain (mb). D) Expression of Pax6.3 at the same stage is seen in the distal part of the optic vesicle and in the posterior diencephalon. E) At stage 22 Pax6.1 expression is seen in the optic cup (oc), diencephalon and hindbrain, while F) Pax6.3 signal is maintained in the posterior half of the optic cup and has increased in the diencephalon. G) A cross section at stage 27 shows Pax6.1 expression in the neuroretina (nr) and lens (l) epithelium of the eye, and in the dorsal diencephalon. H) In contrast Pax6.3 signal is no longer seen in the retina, but strong expression is maintained in the dorsal diencephalon. I) By stage 30 medaka Pax6.1 expression is restricted to the lens epithelium, the ganglion cell layer and the inner nuclear layer in the retina, and in the dorsal and medial diencephalon and ventral nerve tracts. K) Pax6.3 expression is limited to the dorsal and medial diencephalon.