- Title
-
The neuroepithelial basement membrane serves as a boundary and a substrate for neuron migration in the zebrafish hindbrain
- Authors
- Grant, P.K., and Moens, C.B.
- Source
- Full text @ Neural Dev.
|
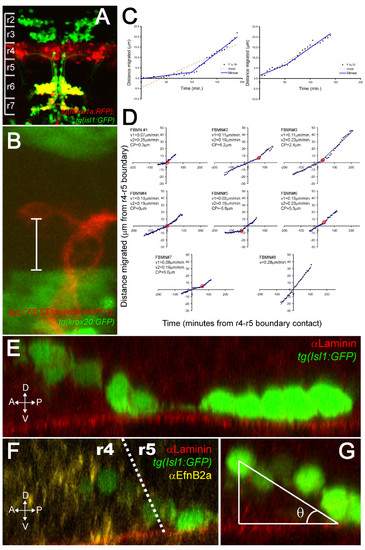
FBMNs undergo a change in velocity upon contact with the Laminin-containing ventral basement membrane. (A) A maximum intensity projection of a confocal Z-stack of a tg(hoxb1aBAC:mRFP1); tg(isl1:GFP) double transgenic embryo at 48 hpf reveals that most Isl1:GFP positive neurons in both r6 and r7 are also hoxb1a:RFP positive, indicating that they were born in r4. (B) A single XY section of a tg(zCREST1:memb-mRFP1);tg(krox20BAC:GFP) double transgenic embryo at approximately 24 hpf showing the measurement from the posteriormost extent of the FBMN cell body to the r4-r5 boundary (white bar). (C) Plots of the extent of migration of single FBMNs versus time elapsed generated from timelapse movies of a tg(zCREST1:memb-mRFP1);tg(krox20BAC:GFP) double transgenic embryo starting at approximately 24 hpf. In the left graph, points are best fit by a bilinear model rather than a single line, indicating that the FBMN undergoes a single change in velocity while the right graph is equally well fit by either model, indicating a constant velocity. (D) Plots of extent of migration versus time re-centered so that the origin of the graph corresponds to the point of contact with the r4-r5 boundary (negative timepoints and distances indicate migration through r4 while positive timepoints and distances indicate migration through r5). FBMN#1 to FBMN#7 are those best fit by a bilinear model while FBMN#8 is an example of one best fit by a single line. CP, changepoint, the point at which the slope changes in the bilinear model (red circle); v1, initial slope; v2, slope after changepoint. (E-G) Reconstructed YZ sections through 24-hpf isl1:GFP embryos stained for Laminin (E) or Laminin (red) and α Efnb2a (yellow) (F) reveal that FBMNs undergo a ventral migration in r4 and come into close apposition with the basement membrane just after crossing the r4-r5 boundary, after which point their migration is purely posterior. (G) The angle of the ventroposterior early migration with respect to the basement membrane (θ) is measured by defining a right triangle with vertices at the middle of the anteriormost cell, the point of contact with the basement membrane and the basement membrane as one of the legs. |
|
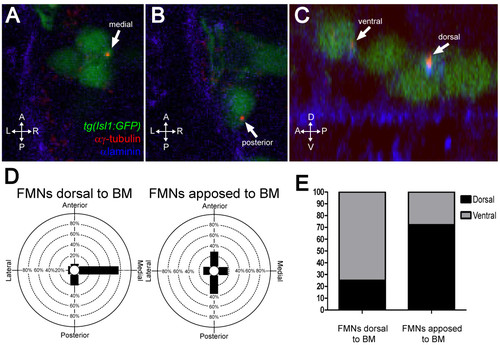
FBMNs undergo a change in centrosome orientation upon contact with the Laminin-containing ventral basement membrane. (A, B) Single XY sections of 24-hpf embryos stained for Laminin (blue) and γ-tubulin (red) reveal that FBMNs that have not yet contacted the basement membrane (BM) (A) mostly orient their centrosomes medially (arrow) while FBMNs that are apposed to the BM (B) orient their centrosomes anteriorly or posteriorly (arrow). (D) Percentage of FBMNs dorsal to the BM whose centrosomes are found in the specified quadrant versus FBMNs apposed to the BM. (C) Reconstructed YZ sections of these same embryos reveal that centrosomes in FBMNs that are dorsal to the basement membrane are mostly in the ventral half of the cell while in FBMNs that are apposed to the basement membrane the centrosomes are mostly dorsal (arrows). (E) Percentage of FBMNs dorsal to the BM whose centrosomes are found in the dorsal or ventral halves versus FBMNs apposed to the BM. |
|
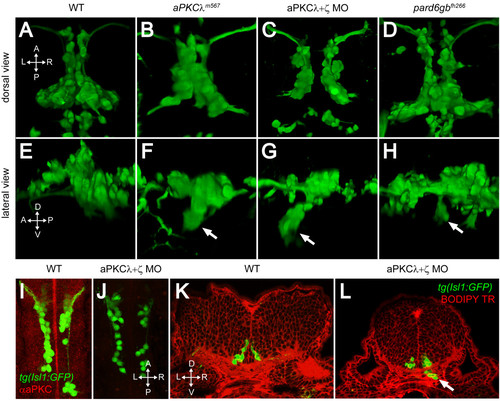
PAR-aPKC complex disruption causes a ventral mismigration in which FBMNs escape the hindbrain ventrally. (A-H) Three-dimensional reconstructions of confocal Z-stacks of the Isl1:GFP positive facial motor neurons at 48 hpf in wild-type (WT) (A, E), aPKCλm567 homozygous mutant (B, F), aPKCλ+ζ double morpholino knockdown (C, G), and pard6gbfh266 homozygous mutant embryos (D, H) shown in dorsal (A-D) and lateral (E-H) views. In aPKCλm567, aPKCλ+ζ double morpholino knockdown, and pard6gbfh266 embryos (F, G, H), a subset of FBMNs mismigrates ventrally (arrows) forming ectopic ventral clusters. (I, J) Maximum intensity projections of confocal Z-stacks of 24-hpf WT (I) and aPKCλ+ζ MO (J) embryos stained with an antibody that recognizes both aPKCs reveals that double morpholino knockdown reduces aPKC to undetectable levels. (K, L) Vibratome coronal sections through 48-hpf WT (K) and aPKCλ+ζ MO (L) embryos at 48 hpf counterstained with BODIPY TR methylester dye that stains all membranes reveals that morphant FBMNs are found outside the hindbrain (arrow). In addition, the cross-sectional area of the aPKCλ+ζ MO hindbrain is reduced and the ventricle has not inflated. |
|
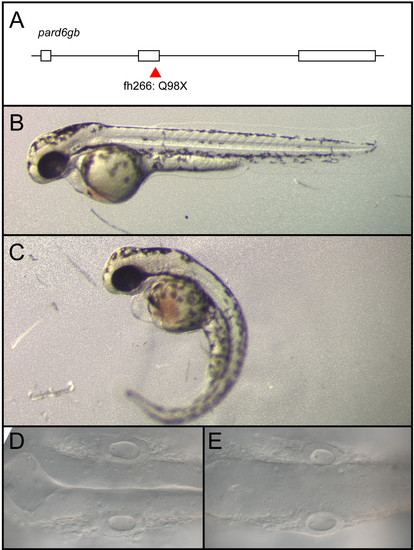
pard6gbfh266 is a nonsense allele that results in edema, curved body axis, and a failure of the hindbrain ventricle to inflate. (A-E) An N-ethyl-N-nitrosourea (ENU)-induced nonsense mutation at the end of exon 2 of pard6gb, pard6gbfh266 (A) results in morphological defects at 48 hpf that include cardiac edema, a curved body axis (C), and a failure of the hindbrain ventricle to inflate (E) when compared to wild type (B, D). These defects are similar to those previously reported for a different allele of pard6gb. |
|
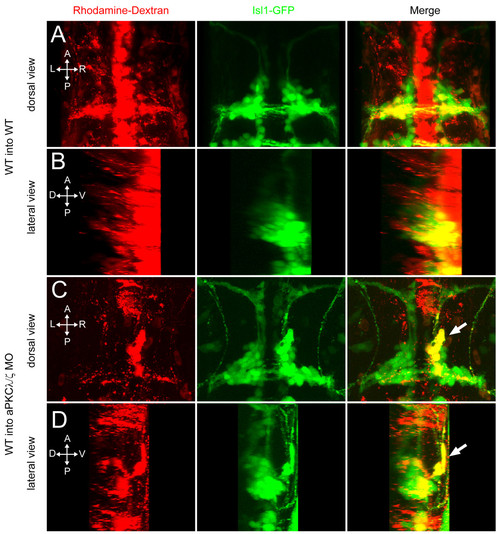
aPKC functions cell non-autonomously to prevent ventral mismigration. Maximum intensity projections of 48-hpf embryos in which rhodamine-dextran-labeled, tg(isl1:GFP) donor cells were transplanted into tg(isl1:GFP) transgenic hosts. (A, B) Yellow colocalization indicates donor-derived FBMNs. Wild-type (WT) donor cells transplanted into WT hosts never mismigrate ventrally. (C, D) A subset of WT FBMNs transplanted into an aPKCλ+ζ morphant host mismigrates ventrally (arrow). |
|
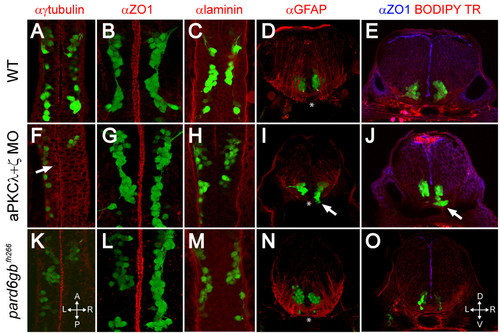
Removal of aPKC or Pard6gb results in defects of apical midline formation and maintenance. (A-C, F-H, K-M) Single XY sections (A, C, F, H, K, M) or three-dimensional reconstructions (B, G, L) of 24 hpf Isl1:GFP transgenic embryos stained for γ-tubulin (A), ZO1 (B), or Laminin (C) show that while apical tight junctions and basal basement membrane are formed correctly in the ventral hindbrain of embryos lacking aPKC or Pard6gb (B, C, G, H, L, M), there are subtle defects in neuroepithelial progenitor polarity reflected in misaligned centrosomes (F) (arrow). (D, I, N) Three-dimensional reconstructions of 70 μm thick 48-hpf cross-sections stained with α GFAP (α-Glial fibrillary acidic protein) show that while staining is reduced in aPKCλ+ζ double knockdown embryos, the absence of radial glial endfeet is unlikely to explain ventral mismigration as endfeet are absent from the most ventromedial region (where mismigration occurs in morphants, arrow) of both wild0type embryos and morphants (asterisks). (E, J, O) Single XY optical sections of 48-hpf vibratome cross-sections stained for ZO1 and with BODIPY TR methylester dye show reduced staining in aPKCλ+ζ double morphants in which ventral mismigration is occurring (J, arrow) or pard6gbfh266 mutants (O), indicating that apical tight junctions are lost between 24 and 48 hpf in these embryos. |
|
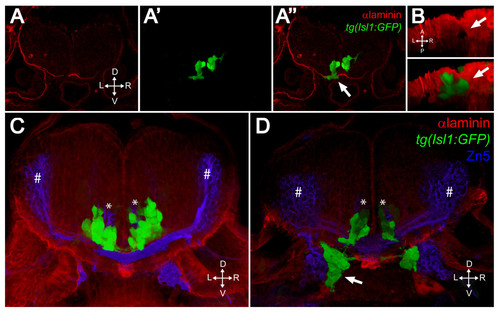
FBMNs exit the hindbrain through holes in the ventral Laminin-containing basement membrane that can be phenocopied by Laminin knockdown. (A-A″) Single XY section of a vibratome cross-section through a 48-hpf aPKCλ+ζ, tg(isl1:GFP) double morphant stained for Laminin (red). Ventrally mismigrating FBMNs exit the hindbrain through a hole in Laminin (arrow). (B) Ventral view of a three-dimensional reconstruction of the same coronal section showing the Laminin hole and the mismigrating FBMNs (arrows). (C, D) Three-dimensional reconstructions of 70 μm thick vibratome cross-sections through 48- hpf wild-type (C) and Lamininα 1 morpholino-injected tg(isl1:GFP) embryos (D) stained for Laminin (red) and Zn5 (blue) showing that Laminin knockdown results in ventrally mismigrated FBMNs (arrow) that exit the hindbrain while leaving abducens motor neurons (asterisks) and commissural interneurons (hash symbols) unaffected. ZN5-staining neurons outside the hindbrain are the sensory neurons of the acoustic nerve (nVIII). |
|
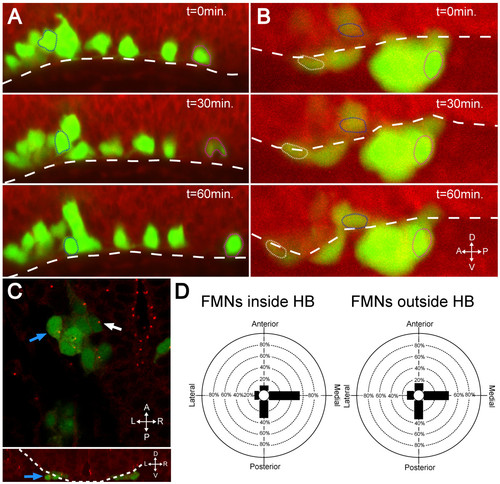
In the absence of Laminin, FBMNs fail to speed up and do not reorient centrosomes. (A, B) Maximum intensity projections from timelapse recordings started at 24 hpf of wild-type (A) and lamininα1-/- (B) tg(isl1:GFP) (green) embryos stained with BODIPY TR methyl ester dye (red) reveal that FBMNs migrate ventrally down to the ventral limit of the hindbrain (dotted line) in both the presence and absence of Laminin (compare blue outlined FBMNs), they do not speed up in the absence of Laminin (compare magenta outlined FBMNs) and some even mismigrate anteriorly (white outlined FBMN). (C) lamininα1-/-; tg(isl1:GFP) embryos stained for γ-tubulin show centrosomes pointing medially in FBMNs both inside the hindbrain (white arrow) and outside the hindbrain (blue arrows). The upper panel is an XY section while lower panel is a reconstructed YZ section at the level of the FBMN indicated by the blue arrow in the upper panel with the hindbrain outlined (dotted line) to show that the indicated FBMN is outside the hindbrain. (D) Quantification of centrosome positioning in lamininα1-/- embryos shows that centrosomes fail to reorient in the absence of Laminin (compare to Figure 2D). HB, hindbrain. |