- Title
-
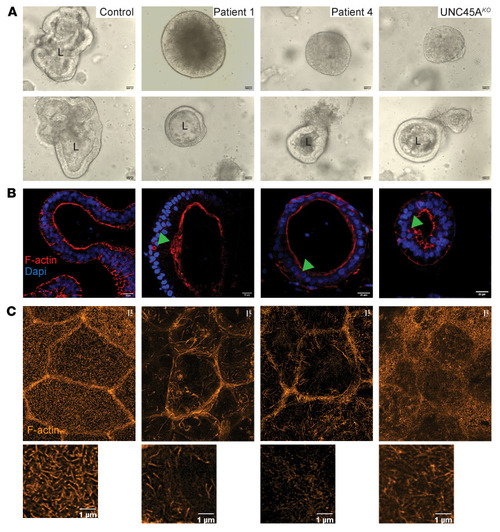
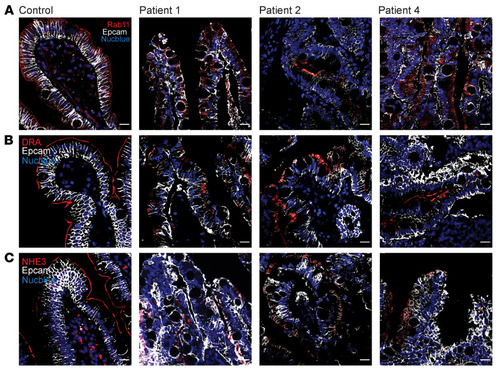
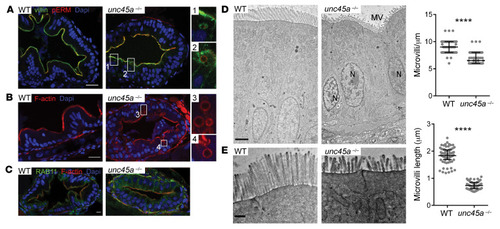
UNC45A deficiency causes microvillus inclusion disease-like phenotype by impairing myosin VB-dependent apical trafficking
- Authors
- Duclaux-Loras, R., Lebreton, C., Berthelet, J., Charbit-Henrion, F., Nicolle, O., Revenu de Courtils, C., Waich, S., Valovka, T., Khiat, A., Rabant, M., Racine, C., Guerrera, I.C., Baptista, J., Mahe, M.M., Hess, M.W., Durel, B., Lefort, N., Banal, C., Parisot, M., Talbotec, C., Lacaille, F., Ecochard-Dugelay, E., Demir, A.M., Vogel, G.F., Faivre, L., Rodrigues, A., Fowler, D., Janecke, A.R., Müller, T., Huber, L.A., Rodrigues-Lima, F., Ruemmele, F.M., Uhlig, H.H., Del Bene, F., Michaux, G., Cerf-Bensussan, N., Parlato, M.
- Source
- Full text @ Journal of Clin. Invest.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |