- Title
-
The endosomal RIN2/Rab5C machinery prevents VEGFR2 degradation to control gene expression and tip cell identity during angiogenesis
- Authors
- Kempers, L., Wakayama, Y., van der Bijl, I., Furumaya, C., De Cuyper, I.M., Jongejan, A., Kat, M., van Stalborch, A.D., van Boxtel, A.L., Hubert, M., Geerts, D., van Buul, J.D., de Korte, D., Herzog, W., Margadant, C.
- Source
- Full text @ Angiogenesis
|
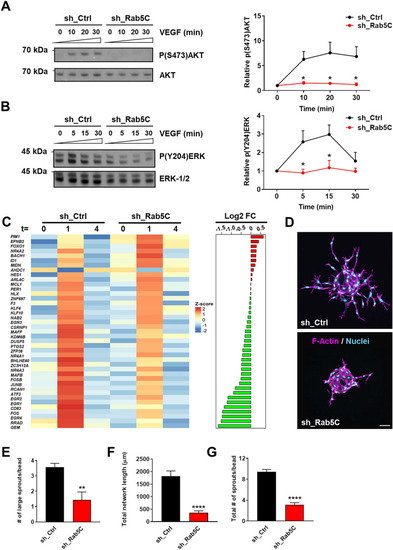
Rab5C protects endosomal VEGFR2 from VEGF-induced lysosomal degradation. |
|
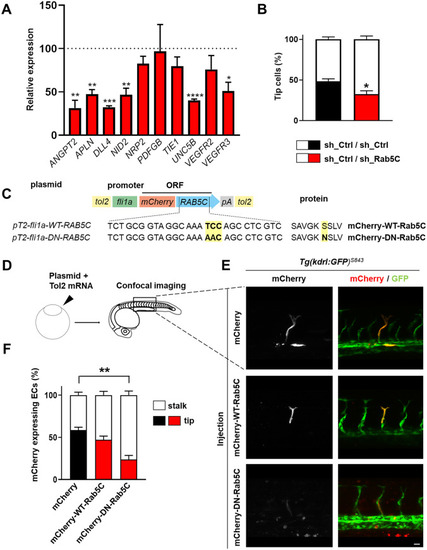
Rab5C regulates endosomal VEGF signaling, the immediate VEGF transcriptome, and sprouting angiogenesis. |
|
Rab5C is essential for tip cell formation. |
|
Rab5C is required for sprouting angiogenesis in vivo. |
|
Rab5C controls Vegf and Notch signaling in vivo. |
|
The Rab5 GEF RIN2 protects VEGFR2 from degradation and promotes angiogenic sprouting. |
|
RIN2 regulates Rab5C recruitment and is required for sprouting angiogenesis in vivo. a Representative Western blot showing the expression of RIN2, endogenous Rab5C, and mScarlet-CA-Rab5C in HUVECs. b Quantification of the total number of sprouts/bead was assessed from confocal microscopy z-stacks for the indicated conditions after 24 h of sprouting. Values represent the means + SEM of n = 33 beads (sh_Ctrl), n = 35 beads (sh_RIN2), n = 48 beads (sh_RIN2 + CA-Rab5C), pooled from 3 independent experiments. c Tg(fli1a:mCherry-WT-hRAB5C)mu227;Tg(kdrl:GFP)s843 zebrafish embryos were injected with control MO (left) or rin2 MO (right) and imaged by confocal microscopy to visualize Rab5C localization. Scale bars, 10 μm. DA, dorsal aorta. d Maximum projections of z-stacks obtained by confocal microscopy showing ISV formation at 32 hpf in Tg(kdrl:GFP)s843 zebrafish embryos injected with Control MO (left) or rin2 MO (right). Scale bars, 40 μm. e Quantification of ISV development (left) as well as average sprout length (right) in zebrafish embryos injected with Control MO or rin2 MO. Control MO: N = 12 embryos, n = 84 ISVs; rin2 MO: N = 10 embryos, n = 72 ISVs. Indicated are statistically significant differences in DLAV and ‘half’ phenotypes compared to Control MO. f Model summarizing the main results of this study. Rab5C and the Rab5 GEF RIN2 protect the EE pool of VEGFR2 from VEGF-induced degradation, which sustains VEGF signaling and is required for the expression of immediate VEGF targets and tip cell genes. Together, these events regulate tip cell specification, endothelial cell migration, and sprouting angiogenesis |