- Title
-
Expression of a Barhl1a reporter in subsets of retinal ganglion cells and commissural neurons of the developing zebrafish brain
- Authors
- Albadri, S., Armant, O., Aljand-Geschwill, T., Del Bene, F., Carl, M., Strähle, U., Poggi, L.
- Source
- Full text @ Sci. Rep.
|
Expression of endogenous |
|
|
|
Time course of |
|
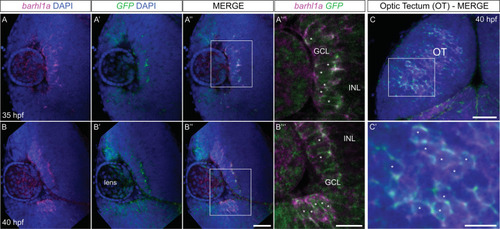
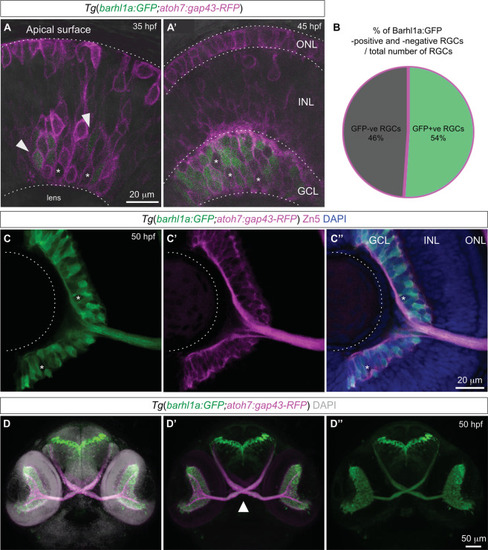
Barhl1a:GFP labels a population of new-born RGCs and their axons. ( |
|
Barhl1a and Ptf1a reporter expressions are mutually exclusive. ( |
|
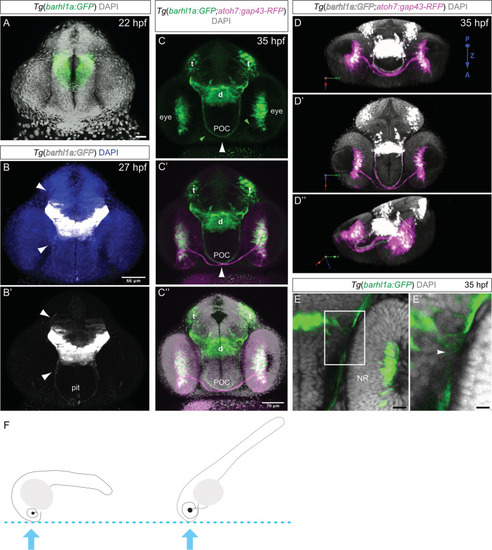
Developmental dynamic of Barhl1a:GFP cells in the diencephalon. Images represent projections of Z-stack ( |
|
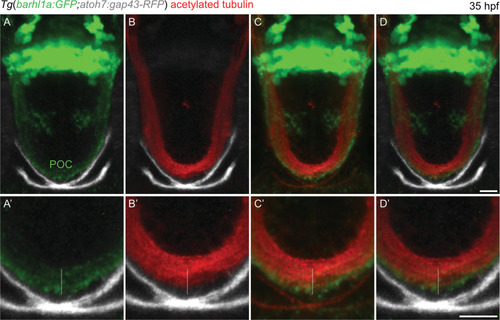
Barhl1a:GFP fibers co-localize with the neuronal projection marker acetylated Tubulin, a marker for the POC. ( |
|
Barhl1a:GFP in the preoptic diencephalon marks populations of mitotic progenitors and differentiating neurons. Images represent single confocal Z-stacks through the diencephalon of |
|
Hypothetical scenario describing Barhl1a RGCs and Barhl2 amacrine cells as clonally related retinal cells forming specific synapses. In this hypothetical model, ( |