- Title
-
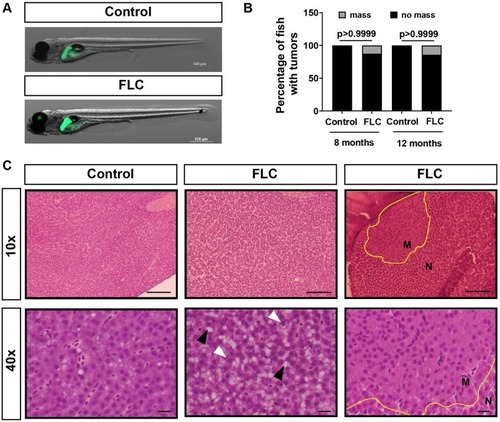
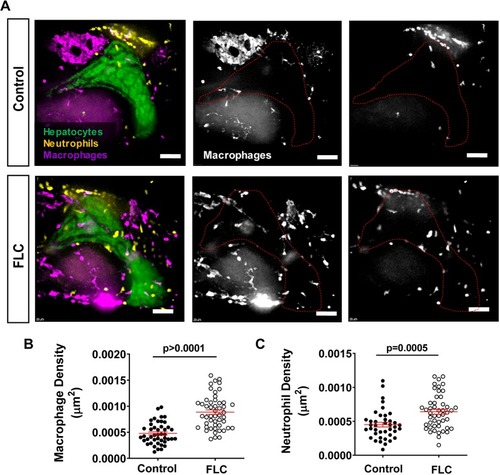
DnaJ-PKAc fusion induces liver inflammation in a zebrafish model of Fibrolamellar Carcinoma
- Authors
- de Oliveira, S., Houseright, R.A., Korte, B.G., Huttenlocher, A.
- Source
- Full text @ Dis. Model. Mech.
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|