- Title
-
Genetic inducible fate mapping in larval zebrafish reveals origins of adult insulin-producing {beta}-cells
- Authors
- Wang, Y., Rovira, M., Yusuff, S., and Parsons, M.J.
- Source
- Full text @ Development
|
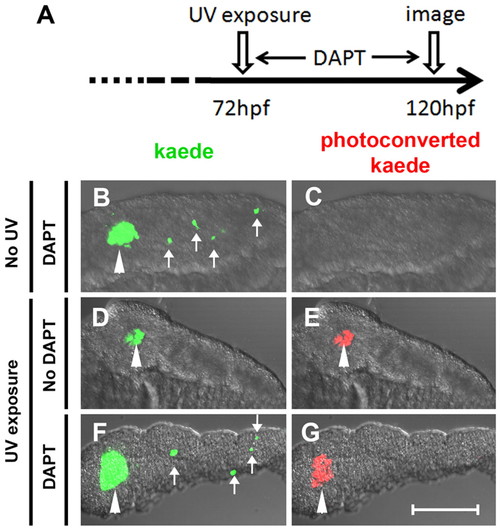
β-Cells of the secondary islets do not originate from principal islet β-cells. (A) Schematic of experimental timeline. The green fluorescent kaede protein in β-cells of ins:kaede fish was photoconverted to red fluorescent protein by UV exposure at 72 hpf. (B-G) Rendered confocal images of microdissected pancreata from ins:kaede fish at 120 hpf. (B,F) DAPT treatment from 72-120 hpf induces secondary islets (white arrows), including differentiated β-cells that are marked by kaede protein. (E,G) Photoconversion at 72 hpf labeled all β-cells present at that time point with red fluorescence. By 120 hpf, photoconverted kaede is still apparent in principal islet cells (white arrowheads) but is not detected in DAPT-induced secondary islets (G). Scale bar: 100 μm. |
|
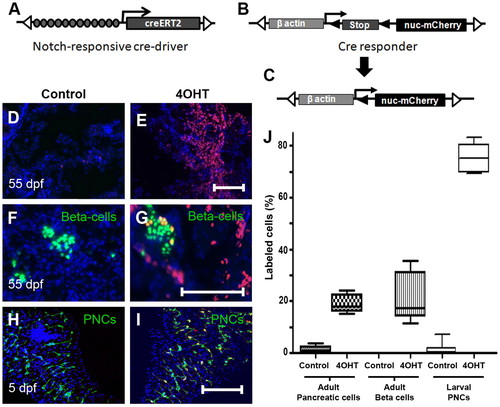
Regulation of Cre-recombinase activity in the pancreas. (A) Schematic of Notch responsive Cre-driver transgene Tp1:creERT2 and (B) the Cre-responder transgene βactin:loxP-stop-loxP-hmgb1-mCherry. (A) Tol2 arms (open triangles) flank 12 RBP-Jκ-binding sites (gray circles), the β-globin minimal promoter (arrow) and the gene encoding the 4OHT-regulated Cre recombinase (CreERT2). (B) The β-actin enhancer/promoter upstream of a loxP-flanked (black triangles) translational stop. (C) Following Cre-dependent recombination, the stop cassette is removed, allowing transcription of a gene encoding nuclear Hmgb1-mCherry (nuc-mCherry). (D-G) Sections through adult pancreas (55 dpf) from GIFM fish. (D) Limited Cre-activity is evident without 4OHT. (E) Treatment with 4OHT at 56 hpf results in widespread labeling in adult (55 dpf) pancreatic tissue. (F,G) β-Cells are marked green in GIFM/ins:hmgb1-eGFP fish. (F) Without larval exposure to 4OHT, β-cells are not labeled with red fluorescent Hmgb1-mCherry. (G) With larval 4OHT treatment (at 56 hpf), adult β-cells are labeled. (H,I) Micro-dissected pancreata from 5 dpf GIFM/Tp1:eGFP larvae. PNCs are marked by GFP fluorescence. (I) 4OHT incubation from 56-80 hpf results in Cre-dependent labeling of 75% PNCs by 5 dpf. DAPI used to counterstain nuclei (D-I). (J) Box plots quantifying results shown in D-I. Scale bars: 100 μm in E,G,I. |
|
Early Notch-responsive progenitors contribute to principal islet β-cells and exocrine cells. (A-P) Confocal images (single optic section) of microdissected pancreata from GIFM larvae at 5 dpf. Fish are also transgenic for either (A-H) ins:hmgb1-eGFP (marking β-cells with nuclear GFP) or (I-P) ptf1a:eGFP (marking acinar cells with GFP). As indicated, larvae were treated either with 4OHT (4OHT treated) or with vehicle alone as a negative control (–ve control; D,H,L,P). 4OHT or vehicle was added at two different stages of development: 12-36 hpf, which is a developmental stage covering the appearance of the pancreas (Early); or 56-80 hpf, a stage of pancreas maturation (Late). (A,E,I,M) Tissue-specific markers in GFP and nuclei in DAPI (blue). (B,F,J,N) Lineage-traced cells with red labeled nuclei. (C,G,K,O) Red and green images merged with bright field. Cells positive for both tissue-specific transgene and lineage labeling appear yellow; examples are indicated with black arrows. Treatment with 4OHT leads to labeling of β-cells of the principal islet and cells of the exocrine pancreas when added at 12 hpf (C,K) but rarely if added later at 56 hpf (G,O). Scale bars: 20 μm in D,H,L,P. |
|
Ventral bud forms from distinct populations of Notch-responsive progenitors and Ptf1a-expressing cells. ptf1a:eGFP; Tp1:hmgb1-mCherry embryo imaged by confocal microscopy at 33 hpf (A,B) and 37 hpf (C). Embryo is positioned at 45° (halfway between a left-side lateral view and a dorsal view) to best observe the pancreas. (A) Bright-field image of fish showing orientation; a, anterior; p, posterior; d, dorsal; v, ventral. Top left-hand side is hindbrain, bottom right-hand side is yolk and positions of the notochord (n), dorsal aorta (da) and intestine (i), are shown. (B,C) Merged fluorescent and bright-field images (single optic sections) at 33 (B) and 37 (C) hpf. Owing to transgenic markers, cells responding to Notch signaling are red (Notch reporter) and cells expressing ptf1a are green (Ptf1a). The lumen of the gut tube is indicated by a broken line and pancreatic Ptf1a expression is detected by green fluorescence (*). PNCs of the ventral bud are in red (arrowhead). (B′,C′) Rendered views of developing pancreas. Optical sections of GFP have been digitally cut away to reveal PNC core of ventral bud in C′. Scale bars: 100 μm. |
|
Pancreatic Notch-responsive progenitors form precocious secondary islets. (A-C,E) Confocal images of single optic sections of micro-dissected pancreata from GIFM; ins:hmgb1-eGFP larvae at 5 dpf. PNCs were labeled by addition of 4OHT from 56 to 72 hpf. All larvae were incubated in DAPT from 72 to 120 hpf, which induces precocious secondary islets. (A) Nuclear GFP expression shows position of β-cells. (B) Nuclear mCherry labels the PNCs and maps their subsequent developmental fate. All nuclear counterstained with DAPI (blue). (C) In red/green images merged with bright-field images, co-labeled cells appear yellow. (A-C) Position of the secondary islet is marked (*) and magnified in the inset panels, where an example of a lineage trace β-cell is marked with a white arrowhead. (D) Schematic of 4OHT-dependent fate mapping and negative control (no 4OHT) in GIFM fish with secondary islet induction. (E) Without 4OHT treatment, no fate mapping is observed. Scale bar: 100 μm |
|
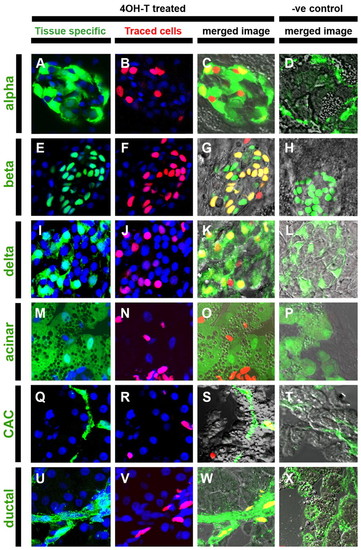
Adult pancreatic endocrine, ductal and centroacinar cells are derived from larval Notch-responsive progenitors. GIFM larvae (56 hpf) were treated with 4OHT (4OHT treated) or vehicle alone (–ve control) for 24 hours and raised to juvenile stages (55 dpf) before pancreata were removed and cryosectioned. (A-X) Confocal images (single optic sections) from sectioned pancreata. (A,E,I,M,Q) Tissue-specific transgenes or (U) immunofluorescence-labeled pancreas cell types in green (tissue specific). Transgenic lines used to mark specific adult tissues: (A-D) gcga:GFP marks α-cells (alpha), (E-H) ins:hmgb1-eGFP marks β-cells (beta), (I-L) SST2:eGFP marks δ-cells (delta), (M-P) ptf1a:eGFP marks acinar tissue (acinar) and (Q-T) Tp1:eGFP marks centroacinar cells (CAC) (note GFP signal in Tp1:eGFP sections is detected by immunofluorescence). (U-X) Immunofluorescent staining was used to detect the ductal marker cytokeratin 18 (ductal). In images of `tissue specific′ marked cells and `traced cells′, nuclei are stained blue with DAPI. Merged images include bright-field views to aid discernment of cellular morphology. (B,F,J,N,R,V) The progeny of fate-mapped PNCs are labeled red (traced cells). (C,G,K,O,S) Fate-mapped PNC progeny contributing to pancreas cells marked by tissue-specific transgene or (W) immunofluorescence result in double-labeled cells that appear yellow in merged images (merged image). Width of each panel represents 50 μm. |
|
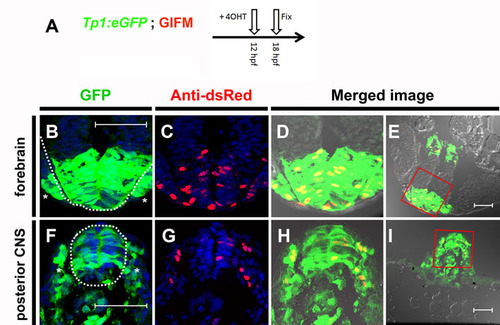
Tp1 Cre-driver restricts recombination in 12 hpf embryos to the Notch-responsive cells. (A) To determine the fidelity of Cre expression in Tp1:creERT2 at 12 hpf, embryos were generated that were transgenic for the GIFM components, as well as for the Notch-responsive marker Tp1:eGFP. As many cells are only transiently Notch responsive, it was necessary to observe co-expression of both eGFP marker and lineage tracer as soon as possible after the addition of 4OHT. As schematized, 4OHT was added at 12 hpf and the embryos were fixed 6 hours later (18 hpf). Six hours is too short a period of time to be able to detect the lineage tracer directly by fluorescence. Instead Hmgb1-mCherry was detected by immunofluorescence using an antibody to dsRed. (B-I) Transverse sections through 4OHT-treated 18 hpf GIFM; Tp1:eGFP embryo. Images are rendered confocal micrographs. Scale bars: 50 μm. (B,F) Expression from the Tp1:eGFP transgene marks Notch-responsive cells in green. (C,G) Immunofluorescent staining for Hmgb1-mCherry marks lineage traced cells in red. Owing to DAPI staining, all nuclei are marked in blue. (D,H) Red and green images merged with bright-field images show all lineage-traced cells are also still Notch-responsive. (E,I) Lower magnification images show orientation for views in B-D,F-H, respectively. (B-E) Section through developing forebrain with the neural epithelium of the telencephalon highlighted with broken white line in B with sensory placodes marked (*). (F-I) Section through posterior of embryo with developing spinal cord of the central nervous system (CNS) highlighted with a broken white line in F and pre-somitic mesoderm marked (*). |
|
Tp1 Cre-driver restricts recombination in 56 hpf embryos to Notch-responsive cells. (A) As schematized, 4OHT was added at 56 hpf and the embryos were fixed 6 hours later (62 hpf). Again, lineage tracer was detected by immunofluorescence using an antibody to dsRed. (B-M) transverse sections through 4OHT-treated 62 hpf GIFM/Tp1:eGFP embryo. Images are rendered confocal micrographs. Scale bars: 50 μm. (B,F,J) Expression from the Tp1:eGFP transgene marks Notch-responsive cells in green. (C,G,K) Immunofluorescent staining for Hmgb1-mCherry marks lineage traced cells in red. Owing to DAPI staining, all nuclei are marked in blue. (D,H,L) Red and green images merged with bright-field images show all lineage-traced cells are also still Notch responsive. (E,I,M) Lower magnification images show orientation for views in B-D,F-H,J-L respectively. (B-E) Image of a section through the retina where Notch activity marks Müller glia cells. (F-I) Section through trunk of an embryo with Notch activity in cells of the developing central nervous system (CNS). (J-M) Images of a section through the trunk of the embryo at the level of the pancreas. The pancreas is outlined by a broken white line in J and the position of the dorsal aorta is highlighted by an asterisk. |
|
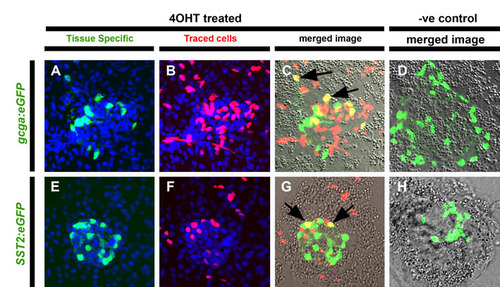
Early Notch-responsive progenitors contribute to principal islet α- and δ-cells. (A-H) Confocal images of single optic sections of micro-dissected pancreata from GIFM larvae at 5 dpf. Fish are also transgenic for either (A-D) gcga:eGFP (marking α-cells with GFP) or (E-H) SST2:eGFP (marking δ-cells with GFP). Larvae were treated either with 4OHT (4OHT treated) or vehicle alone (-ve control) at 12-36 hpf. (A,E) Tissue-specific markers in GFP and nuclei in DAPI (blue). (B,F) Lineage traced cells with red labeled nuclei. (C,G) Red and green images merged with bright field. Cells positive for both tissue-specific transgene and lineage labeling appear yellow and examples are indicated with black arrows. (D,H) Without treatment with 4OHT (negative or -ve control) no endocrine cells are labeled. |
|
Cre-responder line capable of reporting on exocrine fate mapping in 5 dpf larvae. Confocal images of single optic sections of micro-dissected pancreata from β-actin:eGFP-F2A-creERT2; β-actin:loxP-stop-loxP-hmgb1-mCherry 5dpf larvae. (A,B) Following 4OHT addition at 56 hpf, cells within the pancreas parenchyma are labeled with the lineage tracer Hmgb1-mCherry. (A) Counterstaining for nuclei in blue indicates number of cells in field. Red nuclei indicate lineage-traced cells. (B) Merged red and bright-field images show that these labeled or lineage-traced cells include zymogen granule filled acinar cells of the exocrine pancreas. (C,D) Without 4OHT treatment, no labeling is detected. |