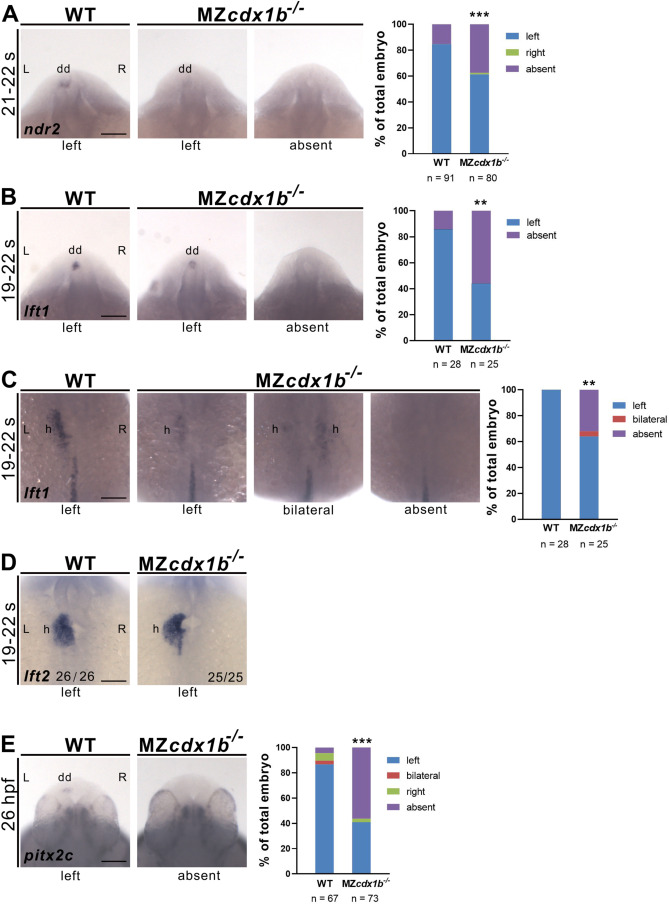
Fig. 3 Fig. 3. The majority of MZcdx1b−/− mutant embryos showed absent expression of ndr2, lft1 and pitx2c in the left dorsal diencephalon and absent expression of lft1 in the heart at segmentation and pharyngula stages. RNA in situ hybridization was conducted on wild-type (WT) or MZcdx1b−/− mutant embryos to evaluate expression patterns in the dorsal diencephalon or heart using ndr2 (A), lft1 (B, C), lft2 (D) or pitx2c (E) RNA probes at 19–22 s or 26 hpf stages. Quantitative data show the percentages of embryos displaying different expression patterns of ndr2, lft1 or pitx2c in the dorsal diencephalon or lft1 expression pattern in the heart in wild-type and MZcdx1b−/− mutants. Chi-squared test of independence was conducted to compare proportions of embryos with absent ndr2, lft1 or pitx2c expression patterns in the dorsal diencephalon or absent expression of lft1 in the heart between wild-type and MZcdx1b−/− mutant embryos. ∗∗, p < 0.01; ∗∗∗, p < 0.001. Scale bar represents 100 μm dd, diencephalon; h, heart, l; left; r, right.
Reprinted from Developmental Biology, 470, Wu, C.S., Lu, Y.F., Liu, Y.H., Huang, C.J., Hwang, S.L., Zebrafish Cdx1b modulates epithalamic asymmetry by regulating ndr2 and lft1 expression, 21-36, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.