Fig. 5
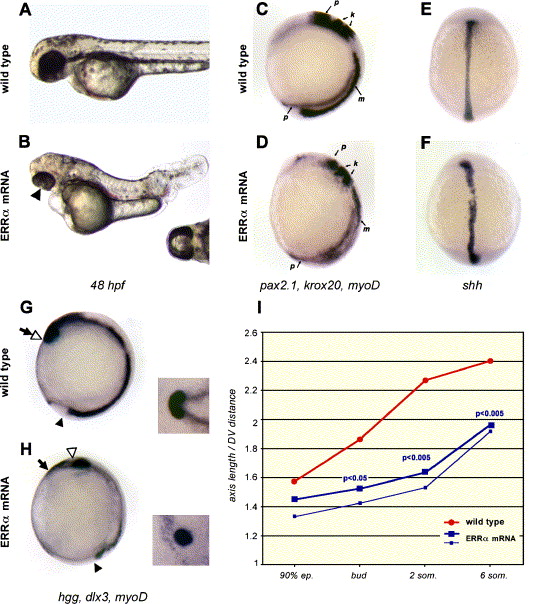
Fig. 5 Axis length reduction in ERRα encoding RNA-injected embryos. (A–B) Lateral views of wild type (A) and ERRα-injected embryos (B) at 48 hpf. Note the tail shortening and the cyclopia of the injected embryo (arrowhead and ventral view of the head in the inset). (C–H) Uninjected (C, E, G) or ERRα mRNA injected (D, F, H) embryos hybridized with a mix of myoD (m), pax 2.1 (p) and krox20 (k) probes (C–D, lateral views of 12 hpf embryos, dorsal to the right), shh probe (E–F, dorsal views of 12 hpf embryos) or hgg, dlx3 and myoD (G–H, lateral views of 11 hpf embryos, dorsal to the right; insets: anterior views of the head). Note that the prechordal plate, labeled by the hgg probe (open arrowhead), has migrated to the tip of the anterior head (delimited by dlx3 expression, arrow) in wild type (G) but not in ERRα mRNA injected (H) embryos. (I) Axis length of wild type (red line) and ERRα mRNA injected (large blue line) from 90% epiboly to 6 somites stages. Axis length is the distance from the tail bud (black arrowhead in panels G–H) to the anterior tip of the head (arrow in panels G–H). P values of the Student′s t test are indicated when a significant difference of the length is measured. Distances between the tail bud and the prechordal plaque (open arrowhead in G–H) have been plotted for ERRα mRNA injected embryos (thin blue line). This distance in wild type embryos is identical to axis length.
Reprinted from Developmental Biology, 281(1), Bardet, P.L., Horard, B., Laudet, V., and Vanacker, J.M., The ERRalpha orphan nuclear receptor controls morphogenetic movements during zebrafish gastrulation, 102-111, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.