Fig. 1
- ID
- ZDB-FIG-250925-1
- Publication
- Olaya et al., 2025 - Distinct cellular and reproductive consequences of meiotic chromosome synapsis defects in syce2 and sycp1 mutant zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
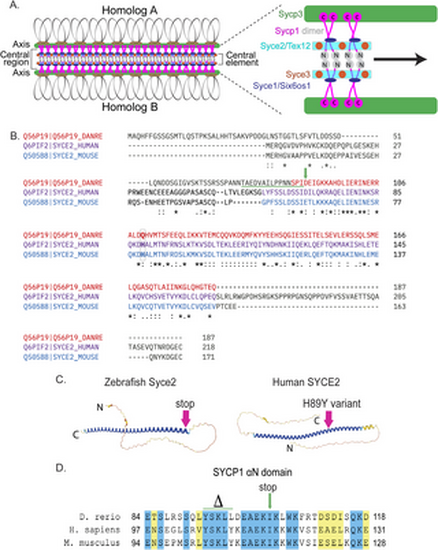
Generating the syce2 and sycp1 mutants. (A) Schematic of the synaptonemal complex showing key structures of meiotic chromosomes: DNA loops (gray/black), chromosome axis (Sycp3, green), central region (Sycp1, magenta) and the central element components (Syce2/Tex12, cyan; Syce1/Six6os1, dark blue; and Syce3, orange) that includes the Sycp1 N-termini (gray). (B) Alignment of the zebrafish Syce2 amino acid sequence with human and mouse orthologs using the Clustal Omega multiple sequence alignment tool [124]. Colored text defines regions that form a long α helix in a 2:2 complex [26]. Conserved identical residues (*), strongly similar (:) and weakly similar (.). The amino acids (aa) affected by the frameshift mutation are underlined in green. The position of the premature stop codon in syce2-/- is shown by a green arrow. The relative position of the human mutation (H89Y) associated with pregnancy loss [6] is highlighted in the boxed region. (C) AlphaFold-predicted structures for Syce2 protein from zebrafish (AF-Q56P19-F1) and human (AF-Q6PIF2-F1) [128]. The position of the 89th codon for zebrafish Syce2 that generates the premature stop codon is shown by a pink arrow. The position of the human variant in SYCE2 is depicted by the pink arrow. (D) Alignment of aa comprising the αN domain of Human SYCP1 that promotes self-assembly [25]. Blue regions are identical aa and yellow regions are similar aa. The CRISPR generated sycp1 mutation is a complex mutation with a deletion that removes the amino acids marked by delta and an insertion that truncates the protein at the amino acid marked by the arrow. See S1 Fig for details. |