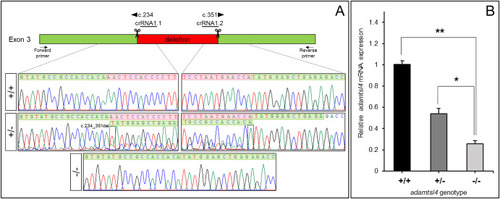
Molecular characterization of the adamtsl4-KO zebrafish line generated using CRISPR/Cas9 genome editing. A. Localization of the deletion identified in adamtsl4 exon 3. Scissors: DNA cleavage sites targeted by two crRNAs (crRNA1.1 and crRNA1.2). The numbers indicate the cDNA nucleotide positions corresponding to the start and end of the deleted DNA fragment. Wild-type sequences are highlighted with a green background, while deleted sequences are marked with a red background. Arrows: position of the forward and reverse primers used for zebrafish genotyping. Rectangles in the electropherograms mark the 5′ and 3′ ends of the deletion. B. Reduced mRNA levels in adamtsl4-KO zebrafish larvae. Relative adamtsl4 mRNA levels were measured in 6 days post-fertilization (dpf) zebrafish larvae using RT-qPCR. Data represent the mean ± standard error, derived from three independent experiments performed in triplicate, using pools of 50 larvae. Statistically significant differences between KO and wild-type zebrafish are denoted by asterisks: p < 0.05 (∗), p < 0.01 (∗∗). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
|

