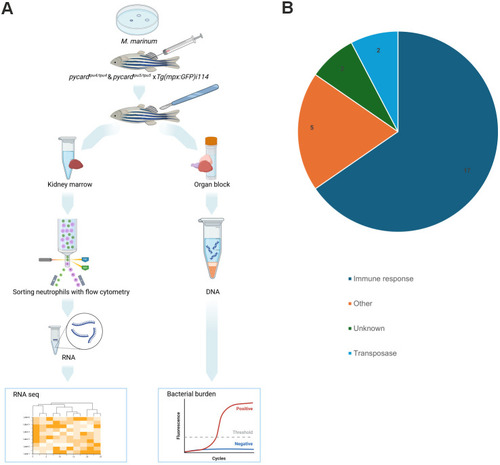
Transcriptomic analysis of kidney marrow-derived neutrophils and categorization of differentially expressed genes in adult pycardtpu4/tpu4 and pycardtpu5/tpu5 zebrafish infected with M. marinum. (A) Fish homozygous for pycardtpu4 and pycardtpu5, and WT controls crossed to Tg(mpx:GFP)i114 (AB) were infected with M. marinum [mean dose, 95 CFU; range, 69-118 CFU (pycardtpu4); mean dose, 6 CFU; range, 4-8 CFU (pycardtpu5)]. At 4 wpi, fish were euthanized and dissected to obtain a kidney marrow and a whole-organ block. From a suspended kidney marrow, fluorescent (GFP) neutrophils were sorted with flow cytometry, and RNA was extracted for transcriptomic analysis with RNA-seq [n(WT)=9, (pycardtpu4/tpu4)=3, (pycardtpu5/tpu5)=6]. The experiment was performed once for pycardtpu4 and pycardtpu5 fish. Both sexes were included in the experiment. To monitor the effect of varying bacterial burden on transcriptomics, bacterial copy number was determined with qPCR from DNA extracted from the whole-organ block. Created in BioRender by Junno, M. (2025). https://BioRender.com/d75w516. This figure was sublicensed under CC-BY 4.0 terms. (B) Differentially expressed genes were categorized according to the literature (Table S4).
|