Fig. 7
- ID
- ZDB-FIG-250219-7
- Publication
- Bouwman et al., 2025 - Cross-species comparison reveals that Hmga1 reduces H3K27me3 levels to promote cardiomyocyte proliferation and cardiac regeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
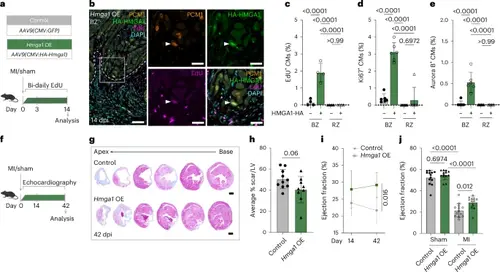
HMGA1 promotes CM proliferation and cardiac regeneration in injured adult mice. a, Schematic overview for experiments in b–e. b, Representative image of immunofluorescent staining against PCM-1, HA and EdU. Dashed line indicates the injury border. Arrowheads indicate HMGA1-HA+EdU+ CMs. Scale bars, 100 μm in overview and 20 μm in zoom-in. c–e, Quantification of EdU+ (c), Ki67+ (d) and Aurora B+ (e) CMs within the BZ and RZ of hearts transduced with HA-HMGA1. n = 4 hearts were analyzed for EdU and n = 6 for Ki67/Aurora B quantification. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show significant differences for % EdU+/Ki67+/AuroraB+ CMs in HMGA1-HA+ BZ CMs compared to HA− BZ CMs and RZ HA+/− CMs (P < 0.0001 for all). No significant difference was found between RZ HA− and HA+ cells for % EdU+ CMs (P > 0.99), % Ki67+ CMs (P = 0.6972) and % AuroraB+ CMs (P > 0.99). f, Workflow for mouse experiments in g–j. g, Representative images of control and Hmga1 OE hearts at 42 dpi stained with Masson’s trichrome. Distance between sections is 400 μm. Scale bars, 1 mm. h, Quantification of scar size in control (n = 10) and Hmga1 OE (n = 9) hearts at 42 dpi showing average % MI length/midline LV length. Error bars indicate mean ± s.d. Statistics were performed by two-tailed unpaired t-test and show no significant difference (P = 0.06). i, Quantification of EF at 14 dpi and 42 dpi of control (n = 13) and Hmga1 OE (n = 13) hearts. Error bars indicate mean ± s.d. Statistics were performed using a two-way ANOVA followed by Sidak’s multiple comparisons test and show a significant difference (P = 0.016). j, Quantification of EF at 42 dpi of sham (n = 13 control, n = 13 Hmga1 OE) and MI (n = 13 control, n = 12 Hmga1 OE) hearts. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show a significant difference between control sham/MI hearts (P < 0.0001), between Hmga1 OE sham/MI hearts (P < 0.0001) and between control and Hmga1 OE MI hearts (P = 0.01) but not between control and Hmga1 OE sham hearts (P = 0.6974). |