Fig. 1
- ID
- ZDB-FIG-250103-23
- Publication
- Herbert et al., 2024 - Identification of a Specific Granular Marker of Zebrafish Eosinophils Enables Development of New Tools for Their Study
- Other Figures
- All Figure Page
- Back to All Figure Page
|
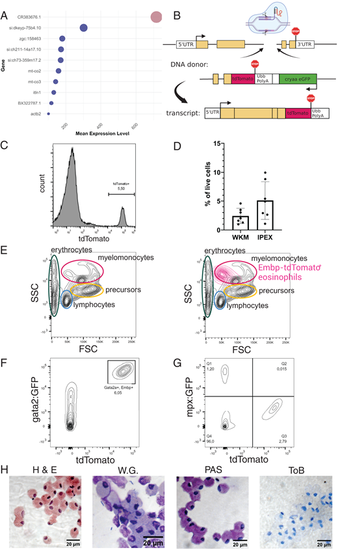
Generation of TgKI(embp-tdTomato,cryaa:EGFP) transgenic reporter line and initial validation. (A) Top 10 most highly expressed transcripts in the eosinophil population of dataset GSE161712 with color-coded information if they are protein coding. Bubble size refers to mean expression levels, and dark blue color indicates protein coding transcripts. (B) Schematic of genetic locus and DNA donor fragment inserted in the embp locus. Transcription leads to a fusion construct of Embp and tdTomato. (C) tdTomato expression in all live, single cells of a representative IPEX sample. (D) Quantification of Embp-tdTomato+ cells in whole kidney marrow (WKM) and i.p. exudate (IPEX) samples of this transgenic line, measured by flow cytometry, and presentation as a proportion of live cells: WKM 2.4 ± 0.38% and IPEX 5.1 ± 2.7% (mean with SD). n = 7. (E) Traditional gating scheme by light-scatter characteristics of a representative WKM sample as previously reported (26). Side-by-side visualization with the Embp-tdTomato+ cell population backgated onto the entire WKM in forward scatter (FSC)/side scatter (SSC). (F) Visualization of Embp-tdTomato and GFP driven by the gata2a promoter in a TgKI(embp-tdTomato,cryaa:EGFP;gata2a:GFP) double transgenic line. n = 3. (G) Visualization of Embp-tdTomato and GFP driven by the neutrophil promoter mpx in a TgKI(embp-tdTomato,cryaa:EGFP; mpx:GFP) double transgenic line. n = 3. (H) FACS-sorted Embp-tdTomato+ cells stained with H&E, Wright–Giemsa (WG), periodic acid–Schiff (PAS), and toluidine blue (ToB). n = 3. |