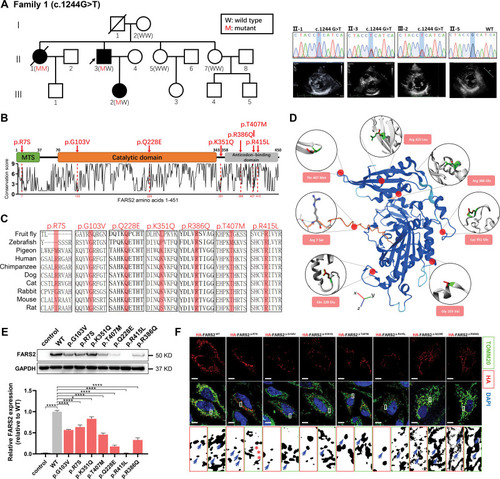
Functional characterization of FARS2 variants identified in hypertrophic cardiomyopathy. A, Pedigrees. Sanger validation of FARS2 variant and echocardiography of the patients in family 1. B, A graphical illustration of FARS2 sequence conservation (bottom) based on ConSurf conservation score. The domain structure and positions of identified patient variants (red symbols) are indicated. C, Species conservation of FARS2 (mitochondrial phenylalanyl-tRNA synthetase) amino acids p.R7, p.G103, p.Q228, p.K351, p.R386, p.T407, and p.R415. D, A structural model of wild-type (WT) human FARS2 (AlphaFold Protein Structure Database O95363 [SYFM_HUMAN]), highlighting the sites of FARS2 variants (p.Arg7Ser, p.Gly103Val, p.Gln228Glu, p.Lys351Gln, p.Arg386Gln, p.Thr407Met, and p.Arg415Leu). E, The expression level of FARS2 from HeLa cells transfected with empty vector control, WT, or 7 variant vectors. The relative statistical analysis is shown in the bottom panel (n=3 per group). F, Confocal images of HeLa cells transfected with HA-FARS2 constructs and immunolabeled with TOMM20 (mitochondrial marker). The regions were magnified with split channels beneath each group and boxed with red (HA) or green (TOMM20). The representative colocalization parts are emphasized by blue arrows; the noncolocalization parts are highlighted by red asterisks. Scale bar=10 μm. *P<0.05; ****P<0.0001.
|