|
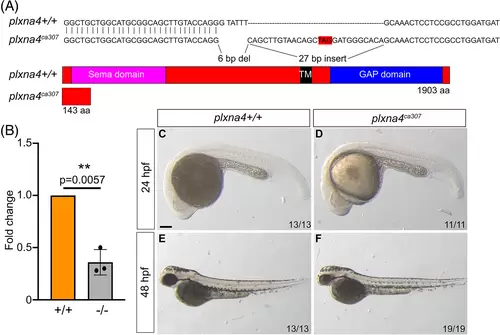
Generation of a CRISPR plxna4 mutant allele: (A) Top: The target nucleotide sequence of exon 1 of plxna4 shows a six base pair (bp) deletion and 27 bp insertion in plxna4ca307 as compared with WT. The alterations in the gene disrupt the WT triplet codon usage and introduce a premature stop (boxed in red). Bottom: The predicted mutant protein is truncated to 143 aa. (B) Fold difference of plxna4 mRNA levels between WT and plxna4ca307 fish as determined by RT-qPCR, with WT data represented as 1. Unpaired Student's t-test performed between the delta CT values for WT and plxna4ca307 fish. N = 3 independent replicates. Error bars are SD. (C–F) plxna4+/+ (C, E) and plxna4ca307 (D, F) embryos at 24 hpf (C, D) and 48 hpf (E, F). Numbers represent combined embryos from two independent clutches. Scale bar: 200 μm (C), 300 μm (E, F).
|