Fig. 4
- ID
- ZDB-FIG-231121-63
- Publication
- Zhang et al., 2023 - Novel biallelic variants in the PLEC gene are associated with severe hearing loss
- Other Figures
- All Figure Page
- Back to All Figure Page
|
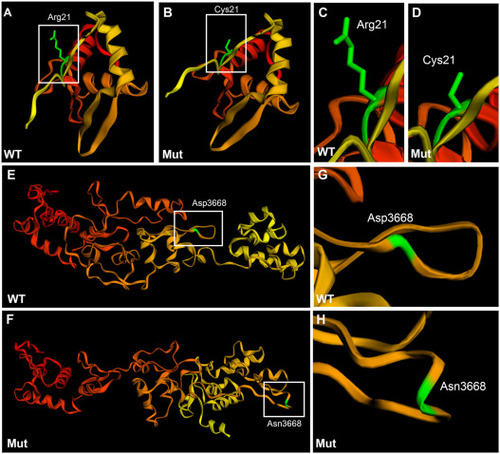
Tertiary structure prediction of wild type and mutant plectin in family #2. A-D: Sequence homology-based prediction approach of the 200 amino acid sequence encompassing the mutation R21C. E-H: Sequence homology-based prediction approach of the 500 amino acid sequence encompassing the mutation D3668N. In the case of the N-terminal region there was no 3D-predicted structure. For the C-terminal region the 3D structure was predicted from the plakin family member, desmoplakin. Phyre2 free tool was used for the analysis. Amino acids are colored code in a gradient from red (N-terminus) to yellow (C-terminus).The amino acid of interest in colored in green. Wild type N-terminus region (A, C), mutant N-terminus region (B, D), wild type 3rd and 4th plectin repeat domains (E, G), and mutant 3rd and 4th repeat domains (F, H). |
Reprinted from Hearing Research, 436, Zhang, T., Xu, Z., Zheng, D., Wang, X., He, J., Zhang, L., Zallocchi, M., Novel biallelic variants in the PLEC gene are associated with severe hearing loss, 108831108831, Copyright (2023) with permission from Elsevier. Full text @ Hear. Res.