Figure 4
- ID
- ZDB-FIG-231002-198
- Publication
- Fernandes et al., 2023 - Complement System Inhibitory Drugs in a Zebrafish (Danio rerio) Model: Computational Modeling
- Other Figures
- All Figure Page
- Back to All Figure Page
|
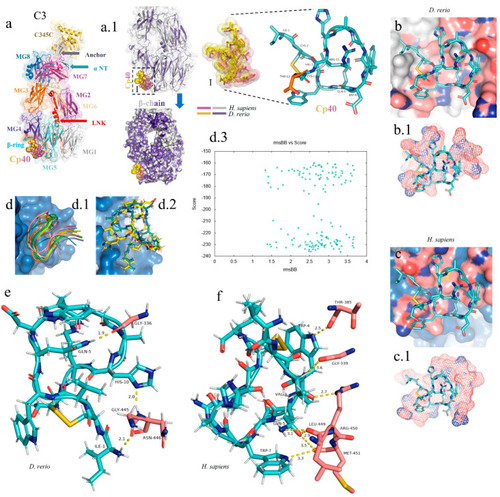
In silico analysis of the peptide–protein interaction between the peptide Cp40 and the C3 molecule of |