Figure 7.
- ID
- ZDB-FIG-230916-237
- Publication
- Nakamae et al., 2023 - DANGER analysis: risk-averse on/off-target assessment for CRISPR editing without a reference genome
- Other Figures
- All Figure Page
- Back to All Figure Page
|
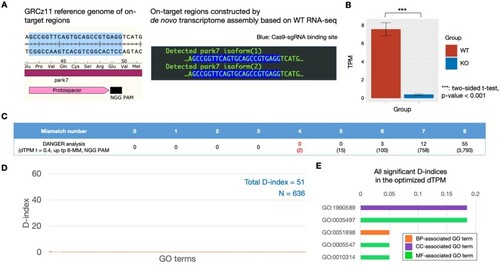
DANGER analysis result using RNA-seq data derived from WT and park7 (dj1) edited brains of |