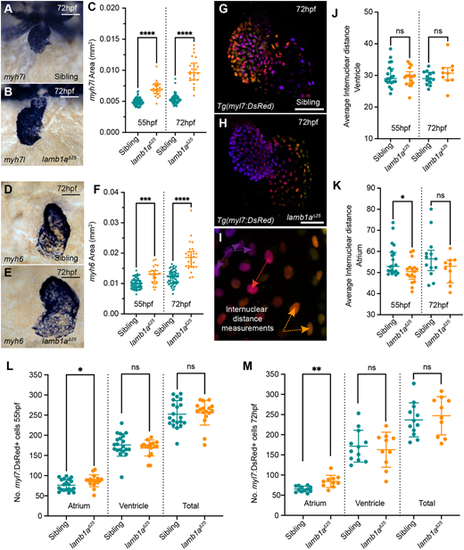
lamb1a mutants have increased atrial cells. (A,B) mRNA in situ hybridisation analysis of myh7l expression in the ventricle of sibling (A) and lamb1aΔ25 mutant embryos (B) at 72 hpf. (C) Quantification of myh7l expression area in sibling (55 hpf: n=72; 72 hpf: n=67) and lamb1aΔ25 mutants (55 hpf: n=23; 72 hpf: n=22). Data are median±interquartile range, analysed with the Kruskal-Wallis test. (D,E) mRNA in situ hybridisation analysis of myh6 expression in the atrium of siblings (D) and lamb1aΔ25 mutants (E) at 72 hpf. (F) Quantification of myh6 expression area in siblings (55 hpf: n=65; 72 hpf: n=64) and lamb1aΔ25 mutants (55 hpf: n=24; 72 hpf: n=29). Data are median±interquartile range, analysed with the Kruskal–Wallis test. (G-I) Depth-coded maximum intensity projections of confocal image z-stacks in Tg(myl7:DsRed) transgenic sibling (G) and lamb1aΔ25 mutants (H) at 72 hpf. Internuclear distance is quantified between nuclei on the same face of the heart that occupy similar z-positions (arrows in I). (J,K) Quantification of average internuclear distance at 55 hpf and 72 hpf in the ventricle (J) and atrium (K) of siblings (55 hpf: n=20; 72 hpf: n=17) and lamb1aΔ25 mutants (55 hpf: n=13; 72 hpf: n=10), demonstrating a mild decrease in the internuclear distance of lamb1aΔ25 mutant atrial cells at 55 hpf (K). Data are median±interquartile range, analysed with Brown–Forsythe and Welch ANOVAs with multiple comparisons. (L,M) Quantification of DsRed+ cells in the myocardium of Tg(myl7:DsRed) transgenic siblings (55 hpf: n=20; 72 hpf: n=17) and lamb1aΔ25 mutants (55 hpf: n=13; 72 hpf: n=10) at 55 hpf (L) and 72 hpf (M). lamb1aΔ25 mutants have a significant increase in atrial cell number at both stages. Data are mean±s.d. Chamber-specific analyses performed with the unpaired t-test with Welch's correction. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05, ns=not significant in all graphs. Scale bars: 50 μm.
|

